Sponsored Content by PittconFeb 3 2017
An interview with Professor David Walt, Tufts University, conducted by April Cashin-Garbutt, MA (Cantab)
Can you please give an introduction to the single molecule array (Simoa) you have developed and that you will be discussing in your talk at Pittcon 2017?
We developed this technology almost 10 years ago. It was developed after a key experiment was carried out by David Rissin, who was a PhD candidate in my laboratory at the time.
At the time, we were working with microwell arrays for a different project. At a lab meeting I posed the question: “How many molecules of a fluorescent molecule would it take to bring the concentration in one of these very tiny microwells to a level that would be easily detectable?” We performed the calculation and it turned out to be about 100 molecules of a fluorescent dye.
David carried out an experiment using a fiber optic microwell array, which had approximately 60,000 microwells on the end of an optical fiber. We trapped a very dilute solution of an enzyme in these microwells–each microwell had a volume of the order of 40 femtoliters (10-15 liters)—a very tiny volume.
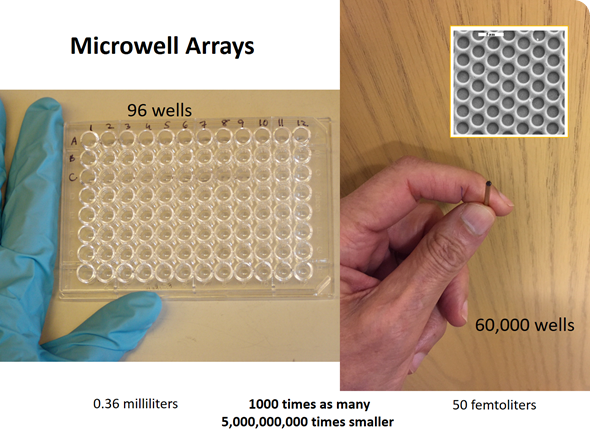
The experiment was designed to trap a very dilute solution of an enzyme containing a high concentration of a substrate. Substrates are the starting materials for enzymes, such that if there were an enzyme present, the enzyme would catalyze the conversion of that substrate into many molecules of a fluorescent product. This process would occur quite rapidly with β-galactosidase, which is a fairly active enzyme.
The idea was that we would run this reaction at such a dilute concentration of enzyme that only a fraction of the wells would contain an enzyme molecule and only those wells that did would fluoresce. After David Rissin worked out the technical details required to carry out the experiment, he demonstrated that one could clearly see which wells contained an enzyme molecule because they turned very bright red in contrast to the wells that contained no enzyme molecule, which remained black.
After this demonstration, we observed that as we decreased the enzyme concentration, we observed fewer fluorescent wells because there were fewer enzyme molecules in the array. We published a paper in Nano Letters and claimed that this was the first time that anyone had measured concentration by counting molecules.
So this experiment was the first demonstration that you could take a solution, confine it into very tiny volumes and literally count the number of molecules present in that solution. In this case, the β-galactosidase concentration could be measured by simply counting the number of bright wells relative to the number of dark wells.
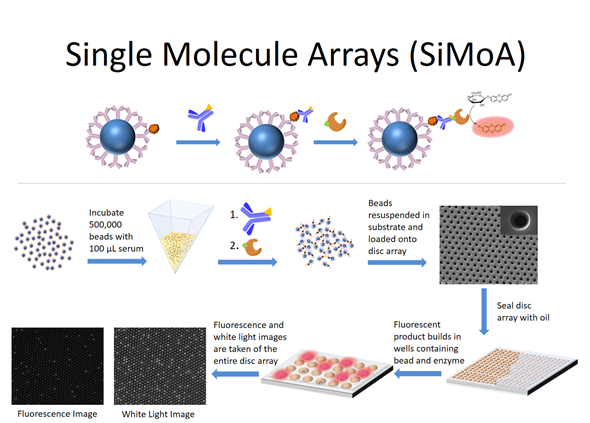
Simoa was developed because we wondered if we could use this approach to actually measure things that people cared about. For example, biologists are particularly interested in measuring molecules such as nucleic acids and proteins.
Over time, the single molecule array technology developed where binding reagents are now attached to microspheres or ‘beads’ in order to capture the target molecules of interest from solution. The beads containing the bound target molecules are then labeled with an enzyme molecule and loaded into the wells and the wells are sealed.
If there is an enzyme molecule attached to a bead, it acts as a reporter for the nucleic acid molecule or the protein molecule of interest. The attached enzyme molecule catalyzes the conversion of many molecules of substrate into product such that wells containing enzyme will build up a locally high concentration of product and fluoresce. The wells containing beads but no bound molecules will stay dark. That's the entire evolution of the technology.
Simoa Technology
How sensitive is Simoa at detecting proteins and nucleic acids?
With respect to nucleic acids, it's close to other sensitive techniques such as the polymerase chain reaction (PCR). For example, there have been demonstrations where a particular nucleic acid sequence can be detected at femtomolar (10-15) concentration so the sensitivity may even be higher than that.
The same goes for proteins where the Simoa limit of detection is typically about a factor of 100 to 1000 times more sensitive than the traditional method of performing a protein assay—the enzyme-linked immunosorbent assay (ELISA) technique. The Simoa technology opens up the potential for being able to measure proteins at concentrations that have never been detected before in various kinds of samples including blood, which is the main focus for Simoa-based assays.
Can you please explain how you have applied the method to the analysis of cultured cancer cells?
We've simply changed the sample handling and preparation so that it can be applied to a single cell assay. It's still a conventional Simoa assay that uses capture antibodies attached to beads in order to bind proteins of interest in individual cells.
For sample preparation, we first isolate individual cells, either manually or with a variety of single cell isolation procedures such as flow cytometry or microfluidics. The key is to isolate individual cells in very small volumes.
We then take the isolated cells and lyse them to disrupt their membranes and release their contents into a small volume of buffer. We then add beads and capture the molecules that we're interested in detecting. Everything then follows the conventional workflow for Simoa.
The goal is to be able to study individual cells within a population. It is now recognized that homogenizing a population of cells, typically thousands if not millions of cells, when making a measurement gives an average concentration of the protein or the nucleic acid that's in the cells. Such measurements mask the heterogeneity of the cell population.
Using Simoa we can measure the protein concentrations in individual cancer cells. Observing hundreds of cells at a time at the single cell level provides insight into the heterogeneity of the cell population. By developing a technology that can measure the protein concentrations in individual cells, we are able to look at the distribution of protein concentrations within the population.
One possible use such a technology would be in a clinical setting to study tissue biopsies. For example, if one took a needle biopsy from a woman who had an abnormal mammogram, the individual cells could then be dissociated from that biopsy tissue and analyzed, as opposed to looking at the population average.
This approach is important because if there were any rare cells in the sample that happened to have a particularly aggressive phenotype, even one out of 1,000 or one out of 10,000 cells in that tissue sample, it would be difficult to detect that cell if you were to homogenize the tissue first and were looking only at the average of the cell population. This particularly rare aggressive cell is going to divide rapidly and could end up leading to metastatic disease and ultimately a poor outcome for the patient.
When pathologists evaluate biopsy samples, they look at the overall cell population and they don't get the level of granularity that's necessary to determine if there is a particularly aggressive rare cell present that will give the patient a bad prognosis. By looking at the average, you might conclude, "Well, 9,999 cells are normal" because that rare 10,000th cell is averaged into the results and the results are therefore biased in favor of the population average.
What other applications of Simoa are there?
There are a wide variety of clinical indications being investigated. This technology has now been commercialized by Quanterix—a company based in Massachusetts of which I am the scientific founder. They licensed the patents that came out of my Tufts laboratory and are commercializing the technology.
My lab has been focused primarily on breast cancer and infectious disease detection. We're now beginning to look at Parkinson's disease using the technology. Other researchers are looking at various inflammatory diseases. There's also a fairly substantial community that's using the technology to look at neurological disorders; in particular, things like traumatic brain injury.
The National Football League in the US has funded some of the research that's looking at markers released from the brain when the brain experiences a traumatic injury. These proteins are released and appear in the bloodstream, and appear to be highly predictive of the extent of the brain trauma.
Other researchers have used the Simoa technology to begin to look at blood markers for Alzheimer's disease. Still others are using Simoa to detect early signs of heart attacks, by measuring certain proteins that are released in the early stages when the heart is beginning to have issues.
What will audience members learn from your talk at Pittcon 2017?
At Pittcon I will be speaking about some of the technical details of sample preparation for getting single cells isolated to make meaningful biological measurements.
I'll also present some of our latest work with various cultured cancer cells where we've been able to multiplex the assays and I will show some meaningful measurements that indicate that we have a good chance of detecting rare cells when we apply this technology to clinical samples.
What Pittcon Can do for You
What Pittcon Can do for You from AZoNetwork on Vimeo.
What impact do you think Simoa will have in the future with regards to detecting rare cells?
I have already mentioned the detection of aggressive rare cells in tissue biopsies in a clinical setting but there's also a research component in addition to the clinical component.
In a research setting, one of the interesting questions from our perspective is what do these rare cells look like? How do they differ from the population average? There's been a lot of effort in single cell genomics as a result of the wide variety of genetic technologies, including sequencing, that are now available. As a result of these technologies, there's the capability to isolate individual cells, tag them and then look at how the genetics differs from cell to cell.
Of course, that is giving us some interesting insight into cell-to-cell variability but one of the things that people are extremely interested in is how do these genetic changes manifest in terms of how the cell expresses different proteins and how the cell actually differs from its surrounding cells?
Again, getting from the genotype to the phenotype is a much less explored area, and is what my lab is focused on—trying to understand how large are these differences between cells. If we can now combine both technologies and correlate the genotype—the genetic differences—with the phenotype—the way proteins are expressed and the physical appearance of the cell—then we're going to have a much better understanding of how rare cells have the ability to do things that most of the other cells in the population cannot do.
Then of course, the next step is to figure out a way to scale up this technology such that it can become a diagnostic technique, so that it can be used in the clinic as well as a research tool.
What do you think the future holds for single molecule detection and single cell analysis?
I think both of these areas are incredibly promising. The thing that unites both single molecule detection and single cell analysis is that the molecule is the fundamental unit of chemistry and the cell is the fundamental unit of biology.
We’re in the very early days of being able to analyze single molecules, look at populations of molecules, and with some of the more biophysical technologies, look at single molecules and observe them over time. My lab has demonstrated this capability with relatively large single molecules such as enzymes.
Single cells are the fundamental units of biology. Just as single cells have differences when you begin to look at the levels of proteins in them or the sequence of DNA in ostensibly identical cells, when you look at single molecules, you also see a distribution of behaviors.
We're now at the stage where we have the technologies and the resolution to begin to observe these kinds of individual differences between the behaviors of individual molecules and individual cells. I think it's going to certainly rewrite some of the textbooks in fields such as biochemistry and biology because there's no such thing as an average molecule or an average cell. Every cell has its own identity. Every molecule has its own identity.
These behaviors have previously been treated as average properties of molecules and cells. The stochastic behavior of individual molecules can have significant implications that affect biochemical processes, pathways and ultimately the properties of individual cells. And you can begin to look at these pathways and processes and surmise how the properties of individual molecules manifest themselves at the single cell level.
What advances in technology would you like to see and why?
One thing that I'd like to continue in my lab is to push the boundary of sensitivity to detect even lower concentrations than we've been able to achieve so far. What we've been able to accomplish so far is to measure proteins at two to three orders of magnitude lower concentration. It would be great to seek out another ten to hundred-fold more sensitivity.
There are a lot more things that we will be able to look at with a higher level of sensitivity. Even today, when we are measuring blood samples, we don't see certain proteins because the Simoa assay technology, even with its thousand-fold higher sensitivity, does not give us the sensitivity that we need to measure some of the molecules present in the blood at lower expression levels. I would like to see continued movement of that lower limit of detection to even more sensitive levels.
I also think that it would be great to be able to increase the level of multiplexing so that more molecules of different types can be measured simultaneously.
Then finally, and this is something that I don't have a technology solution for but certainly something that I'd like to see, I would like to be able to begin to apply this kind of molecular counting technology to small molecules. We use immunoassays and nucleic acid assays that are capturing large molecules, but the technology that we've developed is really not amenable to being able to measure small molecules such as metabolites. Obviously it would be great to be able to do that.
Where can readers find more information?
The field is changing so rapidly that the best thing to do is simply search for “Simoa”, and there will be a hundred articles where the technology's been used for various applications.
About Professor David Walt

David R. Walt is University Professor, Professor of Chemistry, Professor of Genetics, Professor of Biomedical Engineering at Tufts University and is a Howard Hughes Medical Institute Professor. He received a B.S. in chemistry from the University of Michigan and a Ph.D. in chemical biology from SUNY at Stony Brook.
His laboratory pioneered the development of microwell arrays, which resulted in the discovery of bead arrays that revolutionized the field of genetic analysis. Dr. Walt’s laboratory also introduced the idea of digital protein detection by developing a high throughput technology for performing single molecule analysis.
Present efforts in the laboratory are aimed at measuring the variation of protein and gene expression in single cells, fundamental studies of populations of single enzyme molecules and single nanoparticles, as well as the ultrasensitive detection of biomarkers for cancer, infectious disease, and biological threat agents.
Dr. Walt is the Scientific Founder and a former Director of Illumina Inc. and the Scientific Founder and a Director of Quanterix Corp. He is a Director of Cerulean Pharma Inc. and Exicure, Inc. as well as Founder and Director of Ultivue, Inc. and Arbor Biotechnologies.
He previously served as co-chair of the National Academy of Sciences Board on Chemical Sciences and Technology. Dr. Walt has published over 300 peer-reviewed papers and has over 75 issued US patents.
He has received numerous national and international awards and honors for his fundamental and applied work in the field of optical arrays and single molecules including the American Chemical Society (ACS) Kathryn C. Hach Award for Entrepreneurial Success (2017), Ralph N. Adams Award in Bioanalytical Chemistry (2016), Gustavus John Esselen Award (2014), Analytical Chemistry Spectrochemical Analysis Award (2013), the Pittsburgh Analytical Chemistry Award (2013), and the ACS National Award for Creative Invention (2010).
He is a member of the National Academy of Engineering and the National Academy of Medicine, a Fellow of the American Academy of Arts and Sciences, a Fellow of the American Institute for Medical and Biological Engineering, and a Fellow of the National Academy of Inventors.
About Pittcon
 Pittcon® is a registered trademark of The Pittsburgh Conference on Analytical Chemistry and Applied Spectroscopy, a Pennsylvania non-profit organization. Co-sponsored by the Spectroscopy Society of Pittsburgh and the Society for Analytical Chemists of Pittsburgh, Pittcon is the premier annual conference and exposition on laboratory science.
Pittcon® is a registered trademark of The Pittsburgh Conference on Analytical Chemistry and Applied Spectroscopy, a Pennsylvania non-profit organization. Co-sponsored by the Spectroscopy Society of Pittsburgh and the Society for Analytical Chemists of Pittsburgh, Pittcon is the premier annual conference and exposition on laboratory science.
Proceeds from Pittcon fund science education and outreach at all levels, kindergarten through adult. Pittcon donates more than a million dollars a year to provide financial and administrative support for various science outreach activities including science equipment grants, research grants, scholarships and internships for students, awards to teachers and professors, and grants to public science centers, libraries and museums.
Visit pittcon.org for more information.