Since the turn of the century, scientists have been presenting evidence that contradicts previous understanding that all functional proteins have a specific, stable, three-dimensional structure. Research has shown that certain active proteins are at least partly intrinsically disordered in solution.
These intrinsically disordered proteins (IDPs) play a functional role in important cellular processes such as cell signaling, translation, transcription and cell cycle regulation − processes that tend to be disrupted in diseases such as cancer, diabetes, malaria, cardiovascular disease and prion protein related conditions.
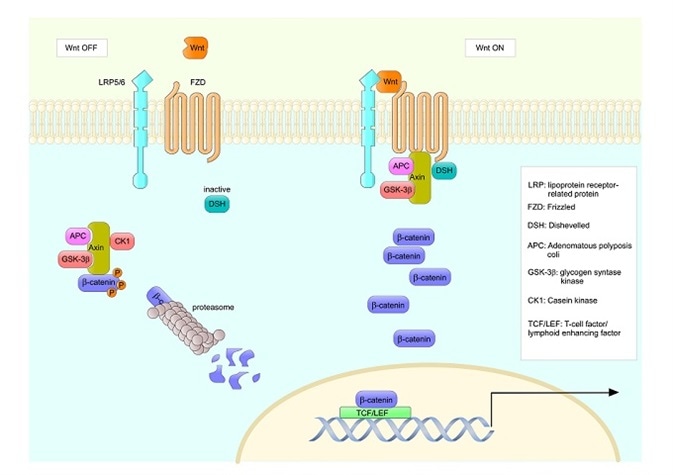
Image Credits: ellepigrafica/shutterstock.com
IDP binding
IDPs often adopt a defined, helical structure when they bind to partner proteins, but intrinsically disordered regions (IDRs) within them can already adopt a helical form, without interacting with a partner molecule. The presence of these regions can significantly influence the protein’s binding process and affinity. Researchers are therefore interested in identifying IDRs with an intrinsic tendency to adopt helical structures in protein segments that interact with binding partners.
Computational characterization
Computational studies are well suited to IDP research, allowing analysis of single molecules at a spatial resolution of up to single atoms. Particularly when combined with techniques such as nuclear magnetic resonance (NMR) spectroscopy, these studies allow atomistic understanding of the various conformations proteins adopt.
One of the main techniques for modeling proteins is molecular dynamics (MD) simulation, which models the motion of atoms by solving Newton’s equations of motion. To do this, the forces that act on the atoms need to be calculated, which requires the use of an analytical water force field model. The force field model is at the heart of any MD simulation.
Most force fields can be relied upon to qualitatively predict the presence or absence of an IDR, but more detailed information on dynamics or sub-states can vary widely depending on the force field used. Researchers have tried to tackle this problem and modify water models, some of which are specifically designed for the description of IDPs.
Researchers from the Technical University of Munich, Germany, recently combined NMR spectroscopy and MD simulation to analyze an IDP called Axin-1. Rainer Bomblies and colleagues found that the predictive ability of their results did indeed depend significantly on which force field water model they used.
Axin-1
Axin-1 is an important IDP involved in the Wnt signaling pathway. It assembles the β-catenin complex responsible for trapping and degrading β-catenin, which regulates cell-to-cell adhesion and gene transcription. Mutations in the gene that encodes Axin-1 have been associated with various cancers including colorectal cancer, hepatocellular carcinoma and endometroid adenocarcinoma.
On forming a complex with β-catenin, Axin-1 adopts a helical structure in the binding region of its partner protein. Researchers predict that the intermediate region of axin-1 (residues 430 to 450) that connects the binding sites in this complex is already in a coiled state prior to binding.
Predicting helicity in Axin-1
Using Bruker’s NMR spectrometers in combination with an advanced MD simulation method, Bomblies and colleagues looked at the helix-forming propensity of Axin-1 across residues 380 to 490. Triple resonance backbone assignment experiments were performed and NMR spectra were recorded using Bruker’s Avance III 600 MHz and Avance II 900 MHz spectrometers. The Avance III represents the next generation in the Avance product line and was designed based on a forward-thinking electronics concept that has made it the fastest and most flexible spectrometer available on the market.
Good qualitative agreement between the experiment and the MD simulation demonstrated that protein segments that bind to signaling proteins did adopt partially helical conformations in the absence of their binding partner. The authors say this is an important finding, since even a small preference for adopting helicity can significantly alter the protein’s binding affinity. They suggest their findings could be used as a basis for understanding how to fine-tune the binding properties of IDPs.
However, the researchers also found that the predicted helical content significantly depended on the force field water model selected. Recent models that have been specifically adapted for the analysis of IDRs reduced the predicted helical content and failed to improve agreement with the experiment, particularly a model called TIP4P-D.
Bomblies and team say their study shows that the choice of force field water model is still of critical importance when studying IDPs and advise that models TIP4P-ws and TIP3P seem to be the most suitable for describing residual helical structures in IDP segments.
References
- Bomblies, R et al. Transient helicity in intrinsically disordered Axin-1 studied by NMR spectroscopy and molecular dynamics simulations. PLoS One 2017: https://doi.org/10.1371/journal.pone.0174337
- Martini, Coarse Grain Forcefield for Biomolecules. The force field jungle, 2016. Available at: http://www.cgmartini.nl/index.php
- NCBI Resources. AXIN1.2017: Available at: https://www.ncbi.nlm.nih.gov/gene/8312
- Emppu Salonen Helsinki University of Technology. Introduction to Molecular Dynamics Simulations, 2006.
- Oldfield, C and Dunker, A. Intrinsically disordered proteins and intrinsically disordered protein regions. Annual Review of Biochemistry, 2014;83: 553-584. https://doi.org/10.1146/annurev-biochem-072711-164947
- University of Cambridge. The Clarke Group. Intrinsically Disordered Proteins. Available at: http://www.ch.cam.ac.uk/group/jclarke/intrinsically-disordered-proteins
- Segditsas, S and Tomlinson, I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene, 2006;25:7531-7537. DOI:10.1038/sj.onc.1210059
About Bruker BioSpin - NMR, EPR and Imaging

Bruker BioSpin offers the world's most comprehensive range of NMR and EPR spectroscopy and preclinical research tools. Bruker BioSpin develops, manufactures and supplies technology to research establishments, commercial enterprises and multi-national corporations across countless industries and fields of expertise.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.