Recombinant protein expression is a fundamental technique that underpins clinical diagnostics, drug discovery and screening, vaccine development, and pure research for elucidating mechanisms of disease development and progression.
Despite its importance, the high-throughput production of human proteins that are fully functional and properly folded frequently encounters difficulties.
The folding of proteins is an intricate process that demands a proper aqueous environment, the presence of chaperones, post-translational modifications, and the formation of multimeric structures stabilized by covalent bonds.
Any alteration in the proper sequence of events can lead to incorrect folding of the protein. Misfolded proteins often result in a loss of function. Utilizing misfolded proteins in subsequent assays or interaction studies can lead to the identification of incorrect biomarkers.
The Sengenics KREX technology incorporates the use of the biotin carboxyl carrier protein (BCCP) as a marker for folding and as a solubility enhancer, allowing for the consistent and efficient expression of full-length proteins that are properly folded and functional.
Biotinylation of BCCP-protein fusions can be performed either in vitro or in vivo, making it possible to utilize the highly specific interaction between biotin and streptavidin for capturing the proteins on the surface.
The binding of biotinylated proteins to a surface coated with streptavidin exhibits minimal dissociation, making it an exceptional method for securing proteins to a planar surface. This interaction is ideal for various applications, including protein microarrays, glass microtiter plates, SPR, and bead-based assays.
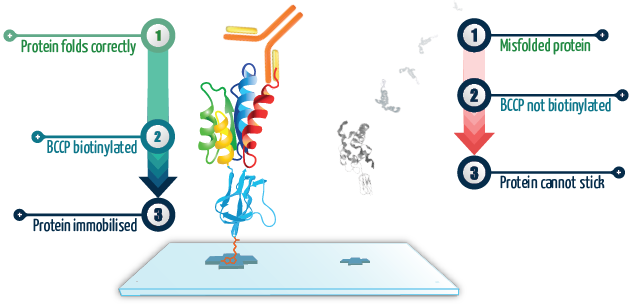
Figure 1. When the protein of interest is correctly folded, it co-translationally drives the correct folding of BCCP, which then becomes biotinylated and allows the fusion protein to become attached to the surface. On the other hand, when the protein of interest is grossly misfolded, it co-translationally drives misfolding of BCCP so it can no longer become biotinylated, preventing it from attaching to the surface. These proteins simply get washed away. Image Credit: Sengenics
Introduction
Besides generating antibodies against foreign substances, the immune system may also produce antibodies against self-antigens, known as "autoantibodies," in response to various pathological processes.
The generation of autoantibodies is thought to occur through mechanisms such as overexpression, mutations, the release of proteins from damaged tissues, misfolding, or incorrect presentation of proteins, causing their recognition by the immune system.
Another advantage of autoantibodies as serological targets is their stability and specificity. They can be conveniently isolated from serum, and their presence can be accurately determined using secondary reagents that have been thoroughly validated.
The immune system's amplification mechanism makes autoantibodies relatively abundant, and the KREX platform allows for their measurement with exceptional sensitivity. This makes autoantibodies an ideal tool for early disease diagnosis.
Citrullination is a modification to the side chains of arginine, resulting from the catalysis of peptidylarginine deiminase (PAD) enzymes. This alteration leads to changes in the protein's structure, antigenicity, and function.
Utilizing the advanced technology of KREX, a fully quantitative protein array platform has been developed. This platform facilitates the simultaneous evaluation of thousands of proteins to determine autoantibody responses concerning various diseases.
The protein array platform is designed such that all the proteins displayed on its surface are correctly folded and functional.
This design enables post-translational modifications, such as citrullination, to be performed on these proteins. As a result, a highly reliable method for identifying autoantibody biomarkers has been developed, particularly in diseases like Rheumatoid Arthritis, Lupus, Multiple Sclerosis, and Alzheimer's.
Citrullination case study
Study design:
The main aim of the study was to discover both previously known and new autoantibody responses to citrullinated proteins by utilizing serum samples from patients with Rheumatoid Arthritis (RA) on the Immunome protein array.
The initial goal was to catalyze the transformation of arginine on the protein surface to citrulline groups on the Sengenics Immunome protein array. To achieve this, the protein array was incubated with peptidylarginine deiminase (PAD) 2 and 4.
Next, the Sengenics i-Ome protein array was subjected to incubation with an anti-citrulline antibody, followed by the addition of a fluorescently labeled detection antibody to identify the presence of citrulline groups.
The secondary aim of the study was to detect known and new autoantibody responses in pooled sera from both RA patients and healthy controls on the citrullinated protein array to identify autoantibodies that are specific to RA.
For this assay, the protein arrays were first incubated with either PAD2 or PAD4 and then subjected to incubation with the pooled sera from RA patients and the matched, healthy control sera. This process was carried out to capture and identify autoantibodies that are specific to RA.
Study objective 1: Detection of citrullinated proteins on the Sengenics i-Ome protein array
Results
The results of the study showed that 70% (1131/1622) of the i-Ome proteins were citrullinated by PAD4, and 73% (1183/1622) were citrullinated by PAD2, displaying significantly greater intensities in comparison to the unmodified Immunome proteins. Additionally, the analysis revealed that 708 citrullinated proteins overlapped between the two types of enzymes.
Table 1. List of 10 out of the 1131 significant citrullinated proteins by PAD4 enzyme on the Sengenics Immunome protein array. Source: Sengenics
| PAD4 Citrullinated Proteins |
| NCK1 |
HUMAN Cytoplasmic protein NCK1 |
| PRKD3 |
HUMAN Serine/threonine-protein kinase D3 |
| PIM2 |
HUMAN Serine/threonine-protein kinase pim-2 |
| AKT1 |
HUMAN RAC-alpha serine/threonine-protein kinase |
| IST1 |
HUMAN IST1 homolog |
| FSCN1 |
HUMAN Fascin |
| TMEM108 |
HUMAN Transmembrane protein 108 |
| TP73 |
HUMAN Tumor protein p73 |
| PXN |
HUMAN Paxillin |
| NCK1 |
HUMAN WNT1-inducible-signaling pathway protein 3 |
Table 2. List of 10 out of the 1183 significant citrullinated proteins by PAD2 enzyme on the Sengenics Immunome protein array. Source: Sengenics
| PAD2 Citrullinated Proteins |
| PRKD3 |
HUMAN Serine/threonine-protein kinase D3 |
| SSB |
HUMAN Lupus La protein |
| AKT1 |
HUMAN RAC-alpha serine/threonine-protein kinase |
| OGG1 |
HUMAN N-glycosylase/DNA lyase |
| VIM |
HUMAN Vimentin |
| PIM2 |
HUMAN Serine/threonine-protein kinase pim-2 |
| NCK1 |
HUMAN Cytoplasmic protein NCK1 |
| HOXA9 |
HUMAN Homeobox protein Hox-A9 |
| HNRNPA2B1 |
HUMAN Heterogeneous nuclear ribonucleoproteins A2/B1 |
| PKM |
HUMAN Pyruvate kinase PKM |
Study objective 2: Identify known and novel autoantibody biomarkers against citrullinated proteins in RA
Results
Citrullination with PAD2: Upon confirming protein citrullination, the PAD2-treated i-Ome protein arrays were subjected to screening using the pooled sera from RA patients and the matched, healthy control sera.
The analysis comparing the two groups revealed increased autoantibody responses toward certain citrullinated proteins that are known to be associated with RA, including SSB, VIM, and HRNRPA2B1 (Figure 3).
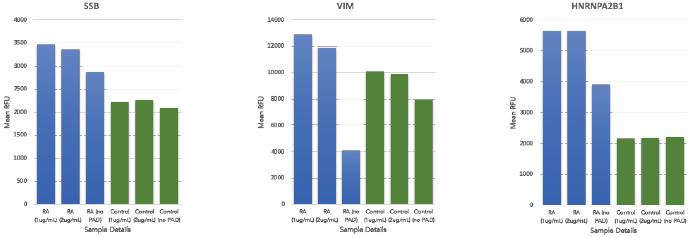
Figure 3. Autoantibody profiles for SSB, VIM and HRNRPA2B1 across all pooled RA (represented in blue) and pooled healthy sera (represented in green) with varying concentrations of PAD2, from left to right: 1 μg/ml, 2 μg/ml and 0 μg/ml. The y-axis represents the average relative fluorescent unit (RFU) between the 4 replica spots of each protein across the different samples. Image Credit: Sengenics
Citrullination with PAD4: After verifying protein citrullination, the PAD4-treated i-Ome protein arrays were screened using the pooled sera from RA patients and the corresponding healthy control sera.
The analysis of the two groups led to the identification of potential new autoantibody biomarkers towards citrullinated proteins in the pooled RA patient sera that were not observed on the non-citrullinated protein arrays (referred to as "no PAD" in the study) (Figure 4).
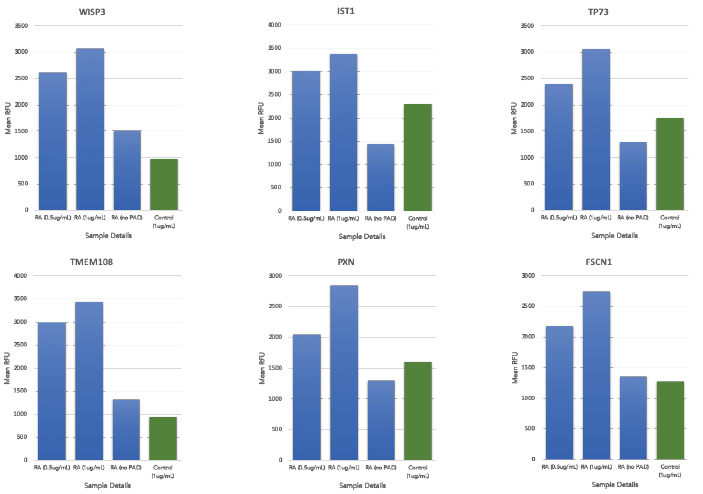
Figure 4. Autoantibody profiles against 6 of the novel citrullinated antigens (WISP3, IST1, TP73, TMEM108, PXN and FSCN1) across all pooled RA (represented in blue) and pooled healthy sera (represented in green) with varying concentrations of PAD4, for pooled RA sera, from left to right: 0.5 μg/ml, 1 μg/ml and 0 μg/ml and for pooled healthy control sera: 1 μg/ml. The y-axis represents the average relative fluorescent unit (RFU) between the 4 replica spots of each protein across the different samples. Image Credit: Sengenics
Conclusion
The ability to post-translationally modify native proteins to citrullinated proteins using PAD2 and PAD4 enzymes has been demonstrated effectively through the use of the Sengenics Immunome protein array platform.
The present study constitutes the most extensive examination of citrullinated proteins to date, involving over 1000 proteins, with a focus on determining the autoantibody profiles in sera collected from patients with Rheumatoid Arthritis.
The stability and specificity of autoantibodies make them particularly advantageous in the context of their role in a broad range of diseases, which are often caused by post-translationally modified proteins such as citrullination, phosphorylation, and oxidation.
About Sengenics
Sengenics is an immunoproteomics company working to improve patient outcomes through physiologically relevant, data-guided decision making. Our solutions enable the discovery and validation of autoantibody biomarker signatures for patient stratification, therapeutic response prediction and elicidation of disease mechanisms.
The company has a global footprint with multiple corporate and research sites across the world with customers and collaborators that include top pharma, biotech and ivy league academic institutions in North America, Europe, and Asia.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.