Recombinant protein expression constitutes a foundational technique that underpins a range of applications, such as clinical diagnostics, pure research, vaccine development and screening, and drug discovery.
However, it is associated with a high rate of failure in the high-throughput production of correctly folded, functional, and full-length human proteins.
This complex protein folding process necessitates an aqueous environment, chaperones, post-translational modifications, and the formation of multimeric structures held together by covalent bonds.
Any deviation from this sequence can result in a misfolded protein, ultimately leading to loss of protein function. The use of misfolded proteins in downstream assays and interaction studies can result in false positives and high background.
To address this limitation, Sengenics KREXTM technology employs Biotin Carboxyl Carrier Protein (BCCP) as a folding marker and solubility enhancer. This technology facilitates the high-throughput expression of full-length, correctly folded, and functional proteins.
BCCP-protein fusions are amenable to being biotinylated in vivo or in vitro, thereby enabling the use of the highly specific biotin-streptavidin interaction for surface capture.
The interaction between biotinylated proteins bound to a streptavidin-coated surface shows negligible dissociation, making it ideal for tethering proteins to a planar surface, thereby providing a vastly superior means for various applications, including protein microarrays, glass microtiter plates, SPR, and bead-based assays.
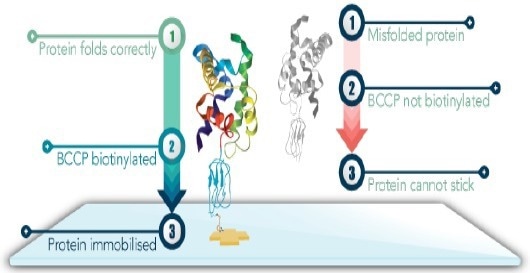
Figure 1. BCCP serves as a folding marker. When the protein of interest folds correctly, BCCP folds correctly and can be biotinyated. This allows it to be imobilized on the streptavadin coated surface. Image Credit: Sengenics
Introduction
DNA alterations, including SNPs, Indels, domain shuffling, or copy number variations, can impact protein properties such as stability, catalytic activity, or the ability to interact with other molecules.
The occurrence of mutations significantly impacts the initiation and progression of several diseases. However, it is challenging to establish a correlation between mutations and protein structure and function, as mutagenesis experiments on physical proteins can be time-consuming and costly.
Advantages
- Access to variant proteins in recombinant form (either purified or over-expressed within a cellular environment)
- Proof that these variant proteins have different functional activity than predicted or recognized
- A faster, more sensitive, and parallel approach for assaying protein variations than current methods
Technical performance
- Expression of correctly folded and functional proteins with a 98% success rate
- Clinical-grade assay metrics provide fully quantitative results with a linear dynamic range of up to five orders of magnitude
- Excellent reproducibility and precision, with a mean CV% below 4% between replica spots
- High sensitivity, with a limit of detection of approximately 1:1,000,000 serum dilution and autoantibody titer of 190 pg/mL
- Exceptional specificity and accuracy, as only correctly folded protins are bound to the array surface
Moreover, only a few methods can evaluate how multiple amino acid substitutions affect a protein's structure and stability. Conventional methods used to assess the impact of genetic mutations on protein function include:
- Newman et al. conducted a comprehensive analysis of yeast sumo structure and function using a high-throughput mutagenic approach. They developed a versatile library comprising over 250 mutant alleles of S. cerevisiae SUMO (Smt3) through the use of biochemical gene synthesis technologies. The library was screened using both traditional colony-forming assays and a high-throughput approach that leverages individual barcodes associated with each mutant allele. As a result, 45 conditional smt3 alleles were identified, enabling the generation of a structure-function map of smt3 and the identification of the residues that are crucial for viability and responses to various cellular stresses.
- Uggenti et al. presented a study that demonstrated the restoration of function for four bestrophin-1 mutant proteins linked to retinopathy to that of the wild-type (WT) protein, using a clinically approved drug, 4PBA. In their previous work, the authors established that nine bestrophin-1 mutant proteins had significantly reduced Cl− conductance compared to the WT protein. They also showed that the expression, localization, and function of the mutant proteins could be rescued through treatment with 4PBA. The authors concluded that 4PBA has the potential to correct protein folding and function in the long term for various diseases.
Utilizing the Sengenics KREXTM technology, we have developed a purpose-designed protein microarray platform, which can facilitate the simultaneous screening of thousands of mutant proteins.
This platform can be leveraged to evaluate the potential of small molecule compounds to restore mutant protein function, providing valuable insights into the development of therapeutics.
This platform enables a superior, low cost, high throughput method for studying the effect of genetic mutations on protein function.
Case study: Parallel characterization of p53 Mutants using protein microarrays
The primary aim of this study is to perform a high-throughput comparison of the effects (e.g., affinity, catalytic efficiency) of naturally occurring p53 variants with SNPs.
A p53 protein microarray was created by cloning and expressing the variants with a downstream His-tag and BCCP-tag, followed by printing on the array surface.
The array was then utilized to evaluate the DNA binding affinity using a cy-3 labeled GADD45 promoter oligo sequence, protein interaction activity of MDM2, and phosphorylation of CKII.
A decrease or complete loss of DNA binding activity was observed in most p53 variants (as shown in Table 1 and Figure 2). However, exceptions were noted for R181C/H, S227T, and H233N/D variants, which contain mutations in solvent that is exposed positions and in regions distal from the protein-DNA interface.
Conversely, R248Q/W, R273C/H, and R280K variants displayed low affinities, indicating a loss of specific protein-DNA interactions due to steric hindrance. Additionally, W23A/G variants demonstrated reduced binding to MDM2, thus indicating a change in protein-protein interaction resulting from the mutation.
Table 1. Partial summary data from microarray experiment. Source: Sengenics
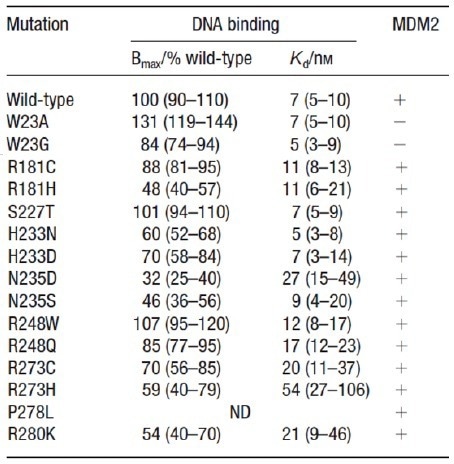

Figure 2. DNA binding of Cy3- labelled GADD45 on p53 custom protein microarray. Image Credit: Sengenics
To determine the functional variation of p53 variants, the researchers performed CKII phosphorylation of p53 at the C-terminal residue S392. Using this method, the p53 variants were phosphorylated with CKII, and the phosphorylation signals were detected with anti-phosphoserine 392 antibodies.
All proteins on the array, except for L344P, 392A (negative control), and variants with mutation X, showed detectable phosphorylation signals.
The presence of premature stop codons introduced at different regions in variants with mutation X resulted in the lack of the S392 region, which is required for CKII interaction, thereby leading to the absence of any detectable phosphorylation signals in these variants.
In conclusion, the reduction in GADD45 and MDM2 binding and the observed lack of phosphorylation signals in p53 variants provide insights into how genetic variation leads to changes or disruptions in protein structure and function.
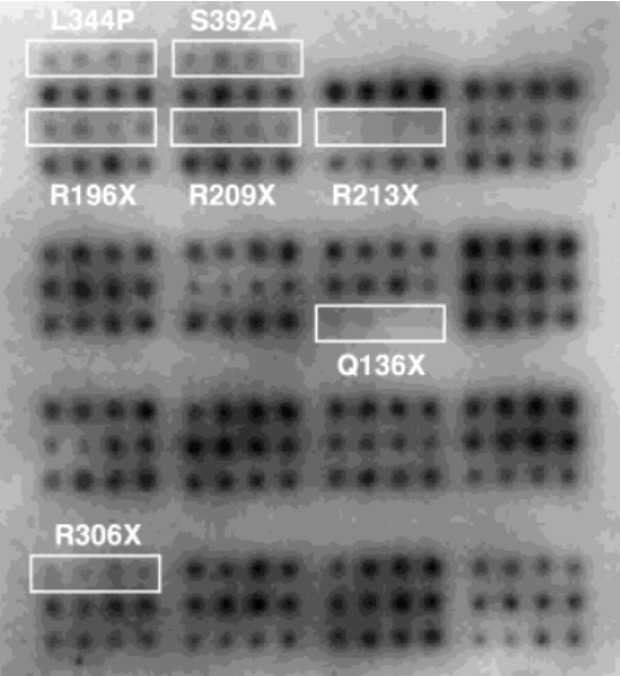
Figure 3. CKII phosphorylation assay on p53 protein microarray. Image Credit: Sengenics
References
- Duarte, J. G. & Blackburn, J. M. (2017) Advances in the development of human protein microarrays. Expert Review of Proteomics 14(7), pp. 627–641
- Boutell, J.M., Darren, J.H., Godber, B.L., Kozlowski, R.Z. & Blackburn, J.B. (2004) Functional protein microarrays for parallel characterisation of p53 mutants. Proteomics, 4, pp. 1950–1958
- 3. Newman HA, Meluh PB, Lu J, Vidal J, Carson C, Lagesse E, et al. 2017. A high throughput mutagenic analysis of yeast sumo structure and function. PLoS Genet 13(2), p. e1006612
- 4. Uggenti C et al. 2016. Restoration of mutant bestrophin-1 expression, localisation and function in a polarised epithelial cell model. Dis Model Mech., 9(11), pp. 1317–1328
About Sengenics
Sengenics is an immunoproteomics company working to improve patient outcomes through physiologically relevant, data-guided decision making. Our solutions enable the discovery and validation of autoantibody biomarker signatures for patient stratification, therapeutic response prediction and elicidation of disease mechanisms.
The company has a global footprint with multiple corporate and research sites across the world with customers and collaborators that include top pharma, biotech and ivy league academic institutions in North America, Europe, and Asia.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.