The expression of recombinant proteins is a crucial technique in various fields, including clinical diagnostics, drug discovery, vaccine development, and research on the mechanisms of disease.
Despite its importance, high-throughput production of full-length human proteins that are correctly folded and functional is challenging.
Protein folding is a complex process that requires a suitable aqueous environment, chaperones, post-translational modifications, and the formation of multimeric structures held together by covalent bonds.
Any deviation from this sequence can lead to misfolded proteins, which can result in false positive biomarkers in downstream assays and interaction studies.
The Sengenics KREX technology overcomes this challenge by using the biotin carboxyl carrier protein (BCCP) as a folding marker and solubility enhancer, enabling consistent high-throughput expression of correctly folded and functional proteins.
BCCP-protein fusions can be biotinylated in vivo or in vitro, allowing specific biotin-streptavidin interactions to be used for surface capture.
As biotinylated proteins bound to a streptavidin-coated surface show negligible dissociation, this interaction provides a robust means of tethering proteins to a planar surface. This is especially useful for applications such as protein microarrays, glass microtiter plates, SPR, and bead-based assays.
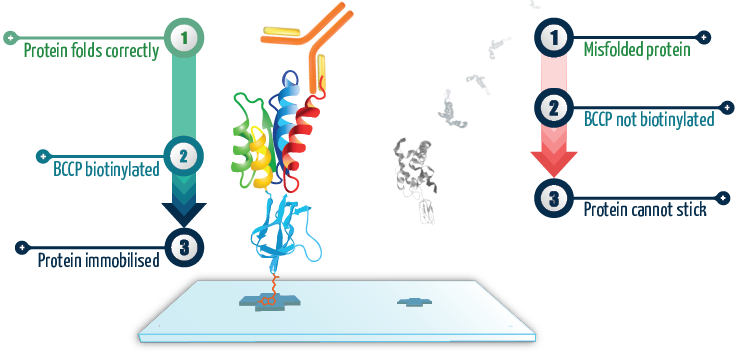
Figure 1. When the protein of interest is correctly folded, it co-translationally drives the correct folding of BCCP, which then becomes biotinylated and allows the fusion protein to become attached to the surface. On the other hand, when the protein of interest is grossly misfolded, it co-translationally drives misfolding of BCCP so it can no longer become biotinylated, preventing it from attaching to the surface. These proteins simply get washed away. Image Credit: Sengenics
Introduction
Viral infections are facilitated by several protein-protein interactions, with protein domains serving as the basic units that define these interactions. Mutations at protein interfaces can alter the binding affinities by modifying the protein's electrostatics and structural properties.
During a viral infection, pathogenic and host cellular proteins engage in constant competition for binding partners. Endogenous interfaces that mediate intraspecific interactions, such as viral-viral or host-host interactions, are persistently targeted and inhibited by exogenous interfaces mediating viral-host interactions.
Blocking these interactions constitutes the primary mechanism behind antiviral therapies (Brito and Pinney, 2017). A protein array that contains antigens from different bacterial or viral strains can be developed to evaluate sera and antibody interactions.
This type of array can allow for a simultaneous examination of the magnitude of antibody responses, the isotype of these antibodies, and the breadth of recognition of different bacterial or viral strains.
Vaccine screening case study
Study design
The onset of Bacterial Meningitis is caused by Neisseria meningitidis, which circumvents the immune system through its Factor H Binding Protein (fHBP) binding with complement factor H.
To discover new vaccines for meningitis, a custom-made Sengenics KREX protein array was created, which contained four different recombinant fHBP variant proteins, each tagged with either N- or C-terminal BCCP.
The array was used to evaluate the binding kinetics of JAR4 and JAR5 meningitis monoclonal antibodies at varying concentrations.
Sera collected from rabbits that were immunized with two protein-based vaccines, Tumenba®-Pfizer and Bexsero®-GSK, were also analyzed on the arrays. A comprehensive description of the samples used in the experiment can be found in the accompanying table.
Table 1. Source: Sengenics
| Name |
Description |
Commercial Source |
| JAR4 |
Anti-Meningococcal factor H binding protein variant 1 (JAR4) monoclonal antibody |
NIBSC |
| JAR5 |
Anti-Meningococcal factor H binding protein variant 1 (JAR5) monoclonal antibody |
NIBSC |
| RS-T |
Serum collected from rabbit immunized with MenB-FHbp Meningococcal Group B Vaccine |
Tumenba®, Pfizer |
| RS-B |
Serum collected from rabbit immunized with MenB-F4C Meningococcal Group B Vaccine |
Bexsero®, GSK |
| IST1 |
Serum collected from rabbit with no immunization (Negative Control) |
Not Applicable |
Results
The kinetic data showed that both JAR4 and JAR5 bind to the N- and C-terminal tagged variant B24 with nanomolar range affinity, which is typical for monoclonal antibodies (as depicted in Figures 2A and 2B).
Tumenba® (made by Pfizer) was found to have a strong affinity to variants B01 and A05, with low-to-mid nanomolar Kd-values, while Bexsero® (made by GSK) had a high affinity to variants B01 and B24, with low nanomolar and micromolar Kd-values, respectively (shown in Figures 2C and 2D).
The negative control sera did not show any binding to any of the fHBP variants (Figure 2E).
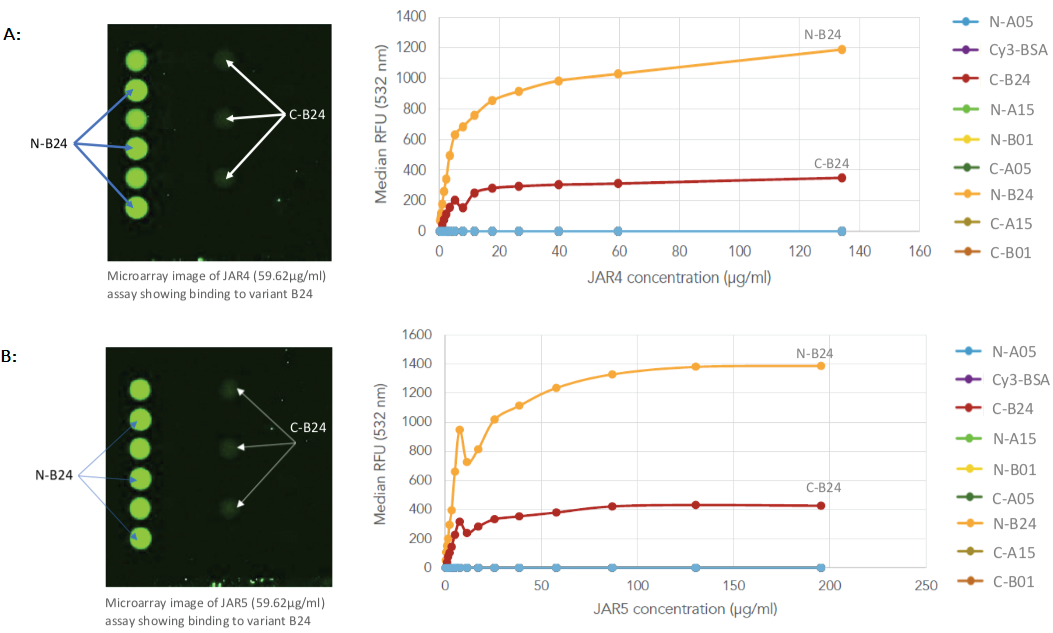
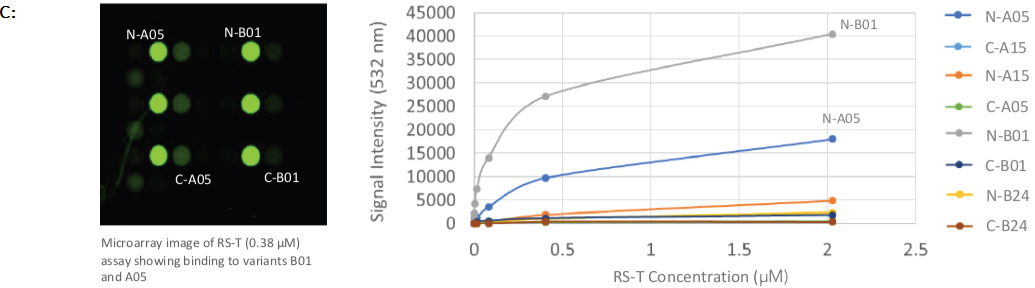
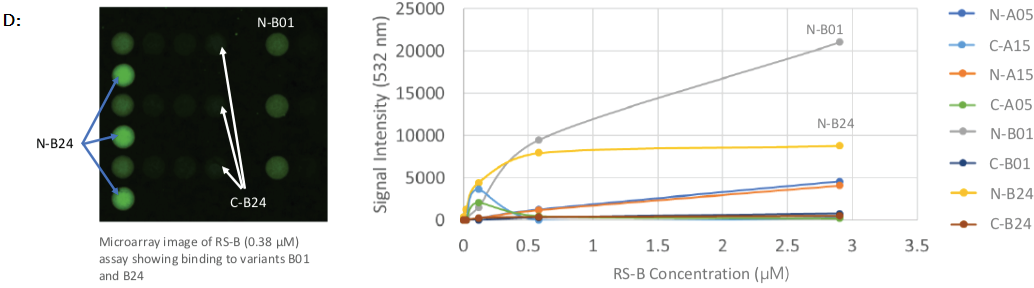
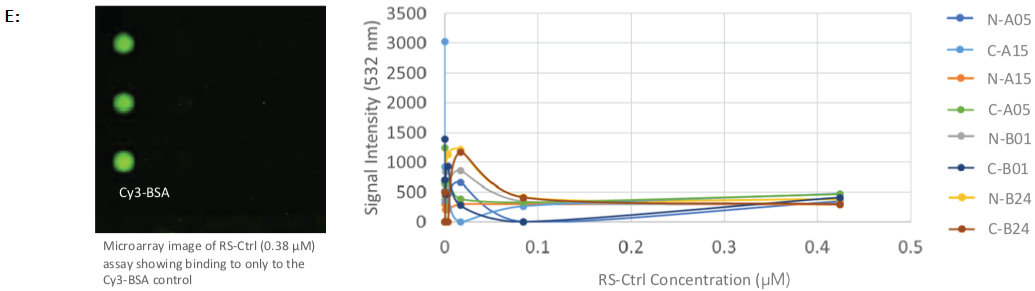
Figure 2. Protein array images and binding curves for all samples assayed across the Sengenics fabricated KREX protein array containing 4 recombinant N- or C-terminally tagged factor H binding proteins (fHBP); A & B: Protein arrays incubated with known meningitis monoclonal antibody vaccines, varying concentrations of JAR4 and JAR5 showed binding towards C- and N-terminally tagged variant B24 only; C: Protein arrays incubated with rabbit serum, RS-T at different concentrations, showed binding towards C- and N-terminally tagged variants A05 and B01; D: Protein arrays incubated with rabbit serum at different concentrations, RS-B, showed binding towards C- and N-terminally tagged variants B01 and B24; E: Protein arrays incubated with negative control rabbit serum at different concentrations, RS-Ctrl, showed no binding towards any of the fHBP variants on the arrays. Image Credit: Sengenics
Conclusion
The study demonstrated the successful application of KREX for the construction of a highly sensitive and specific platform incorporating variations of factor H Binding Protein (fHBP) as a viable tool for screening and finding vaccine candidates against meningococcal disease.
The findings of the study revealed the expected differential binding of the various "vaccine candidates" (monoclonal and polyclonal antibodies in pure and sera forms, respectively) to the various types of fHBP immobilized on the surface.
Moreover, the platform was able to generate precise mechanistic features for each of the "vaccine candidates" probed on the arrays, which is superior to conventional screening approaches.
About Sengenics
Sengenics is an immunoproteomics company working to improve patient outcomes through physiologically relevant, data-guided decision making. Our solutions enable the discovery and validation of autoantibody biomarker signatures for patient stratification, therapeutic response prediction and elucidation of disease mechanisms.
The company has a global footprint with multiple corporate and research sites across the world with customers and collaborators that include top pharma, biotech and ivy league academic institutions in North America, Europe, and Asia.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.