Soon after its emergence in China late December 2019, SARS-CoV-2 spread rapidly all over the world, giving rise to over 22.25 million confirmed cases of SARS-CoV-2 infection and over 783,000 related deaths, as of 20th August 2020. Because of regulatory issues, there was a deficit in diagnostic capacity for COVID-19 in the United States until early March 2020. However, retrospective analysis of COVID cases now suggests the establishment of community transmission in the United States from January – February 2020.
Current study design
In the current study, the researchers carried out whole genome sequencing of 620 clinical SARS-CoV-2 samples, which were collected by the Johns Hopkins Health System between march 11 – 31, 2020, to investigate the initial spread of SARS-CoV-2 in the United States National Capital Region. The number of samples analyzed in the study covered almost 37% of the total cases diagnosed in the region during the given period. In addition, the researchers performed an analysis between genomic sequencing data and disease phenotypes to understand if the viral genotype is correlated with disease severity or viral transmissibility.
Correlation between viral genetic diversity and disease severity
Of total experimental samples, the researchers selected 143 for genomic sequencing, which produced 114 complete SARS-CoV-2 genomes. They employed a tiling amplicon sequencing method to obtain complete viral genomes. While analyzing, they found that samples with a high viral titer, as well as samples collected in the early phase of infection, are more effective in generating complete genomes.
Next, they compared these viral genomic data with disease phenotypes and found multiple entries of SARS-CoV-2 into the region. Importantly, the researchers observed that for many patients, there was no travel history or contact history with infected persons. This indicates that community transmission had already been started in that region in March 2020. However, they did not observe any specific connection between genetic variations of SARS-CoV-2 and disease outcomes in terms of hospitalization or admission to intensive care units (ICUs). This indicates that despite having genetic diversity, there is no correlation between viral genotype and disease severity.
By analyzing genomic sequencing data of clinical samples collected from 40 hospitalized patients and 22 ICU admitted patients, the researchers revealed that the clinical presentation of the disease is not solely associated with viral mutations (diversity). There may be other factors that collectively modulate patient outcomes. This observation was further strengthened by the fact that samples taken from severely affected patients, including those requiring mechanical ventilation, had viruses that belong to all five major phylogenetic clades.
The lower rate of genetic variability of SARS-CoV-2 may be due to the presence of RNA polymerase with proofreading activity. Moreover, there is less evolutionary pressure on SARS-CoV-2 because of the very rapid viral transmission as well as minimal host immunity to counteract the virus. All these factors together may be responsible for limited genetic variations found in SARS-CoV-2 genomes.
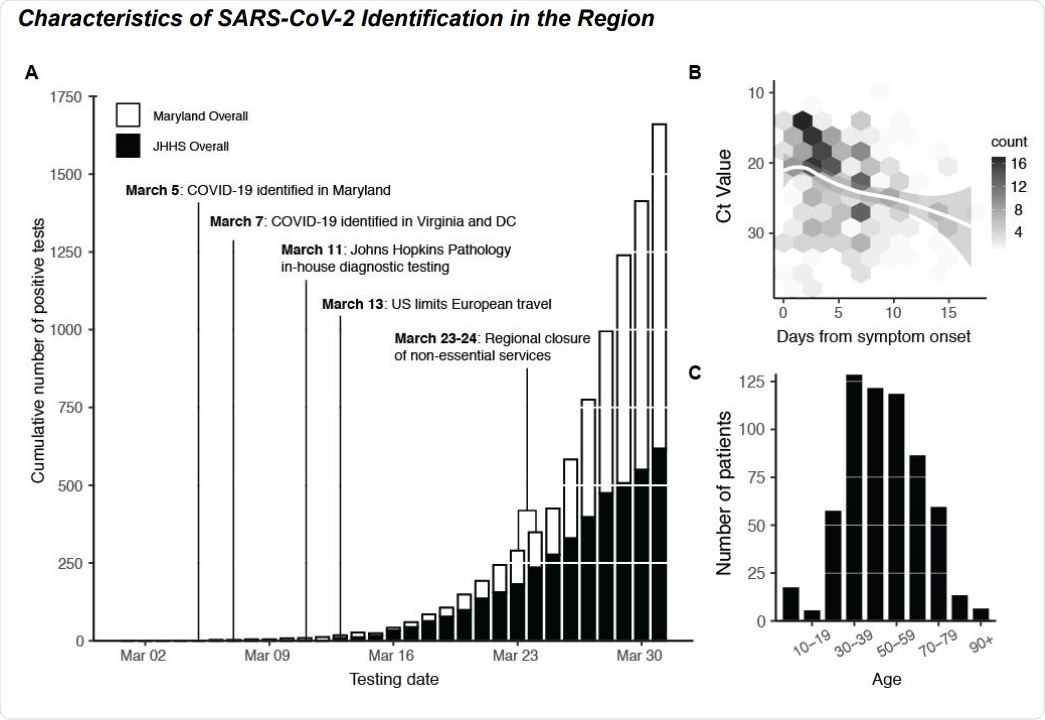
COVID-19 Diagnostic Response During Initial SARS-CoV-2 Surveillance in the Johns Hopkins Health System. (A) Cumulative number of positive tests in the state of Maryland (white bars) and within the Johns Hopkins Health System (JHHS; black bars). (B) SARS-CoV-2 RT-PCR CT value (S-gene) versus days from patient symptom onset. Data fit with LOESS curve (white regression line). Two outliers (days from onset = 5 weeks, CT value = 30 and days from onset = 28 days, CT value = 31) are not shown. (C) Age distribution of SARS-CoV-2 patients within the JHHS.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
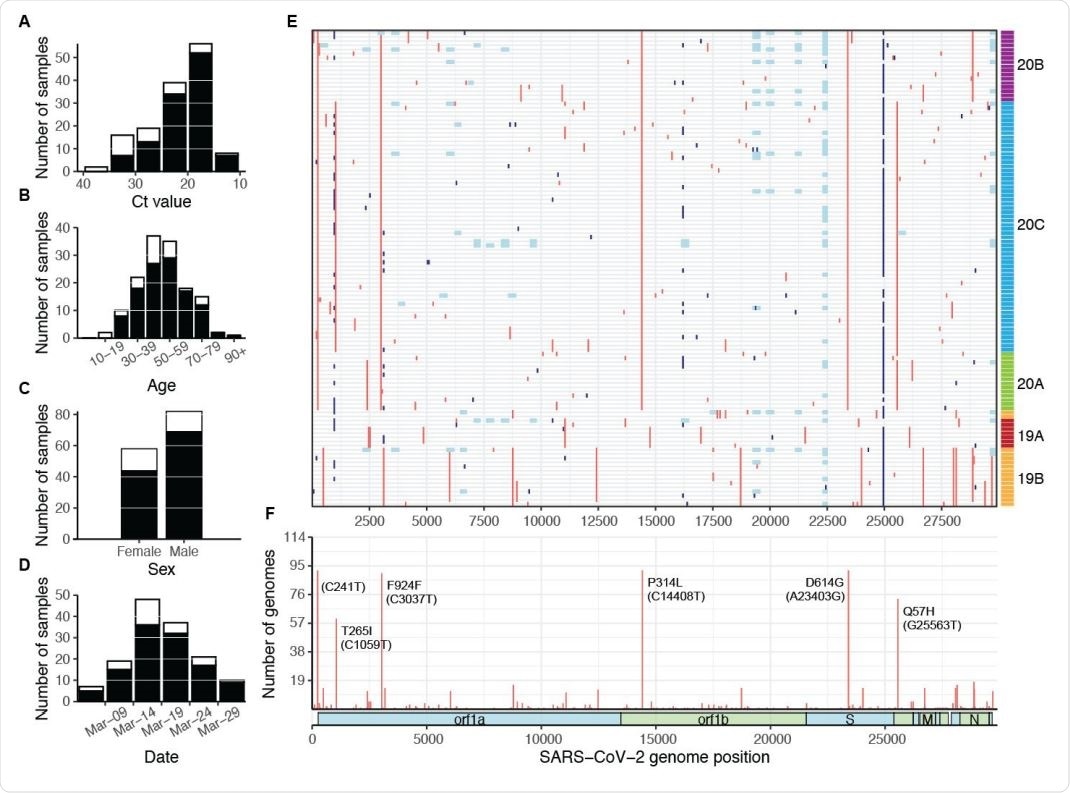
SARS-CoV-2 samples selected for whole-genome sequencing. Distribution of (A) CT value, (B) age, (C) sex, and (D) collection date for specimens selected for whole-genome sequencing (white bars), and specimens that produced complete genomes (black bars). Only specimens with known values are included in each plot. (E) Mutations across the SARS-CoV-2 genome in all 114 complete genomes (rows), binned into 60-nucleotide windows. Red = single nucleotide variant, light blue = base masked as N due to amplicon dropout; dark blue = ambiguous base (N) due to variant-calling issues in homopolymer regions. Rows are clustered by Hamming distance between sequences and colored by phylogenetic clades (19A-20C, see Fig 3). (F) Count of complete genomes (out of 114) with a variant at each site. Key clade-defining mutations are labeled.
Study significance
The researchers believe that the current study findings will allow a better understanding of the SARS-CoV-2 evolution as well as its ever-changing relationship with patient outcomes. In addition, to understand the viral genomic diversity at the initial phase of the COVID-19 pandemic, it is essential to follow these genetic variations and detect corrections between viral genetics and disease severity for ensuring the efficacy of countermeasures (vaccines and therapeutic medicines) that are becoming available to tackle the pandemic.
In the current study, the researchers also compared the viral genomic diversity between different U.S. cities. The data revealed that genetic variations of SARS-CoV-2 observed in New York City seems to be similar to that observed in the National Capital Region because viral entries occurred multiple times in both regions.
Future approach
The researchers are planning to continue their investigation by comparing the genomic sequencing data of viral samples collected during and after the shutdown period. This will help understand the impact of the shutdown as well as the modes of viral spread within this region.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Source:
Journal references:
- Preliminary scientific report.
Thielen, P et al., Genomic Diversity of SARS-CoV-2 During Early Introduction into the United States National Capital Region. medRxiv 2020.08.13.20174136; doi: https://doi.org/10.1101/2020.08.13.20174136
- Peer reviewed and published scientific report.
Thielen, Peter M., Shirlee Wohl, Thomas Mehoke, Srividya Ramakrishnan, Melanie Kirsche, Oluwaseun Falade-Nwulia, Nídia S. Trovão, et al. 2021. “Genomic Diversity of SARS-CoV-2 during Early Introduction into the Baltimore–Washington Metropolitan Area.” JCI Insight 6 (6). https://doi.org/10.1172/jci.insight.144350. https://insight.jci.org/articles/view/144350.