Around five decades ago, Christian Anfinsen carried out advanced experiments which clearly showed that the linear string of amino acids in a protein encodes all the information for its 3D structure. It is now known that this sequence guides the interactions between the solvent and the protein and those between component amino acids. All these interactions are controlled by thermodynamics. However, it is not clear how the sequence encodes the protein’s complex structure. A large number of proteins thermally unfold by 70°C. In proteins, the thermodynamic balance is very delicate. Even a relatively small increase in temperature can denature most proteins. If the balance between unfolded and folded states is maintained by the equivalent of 5 to 8 hydrogen bonds, the question is how are thermophilic proteins stabilized, enabling organisms to thrive at high temperatures and pressures. When there are no thermal changes, it is not clear what thermodynamic events trigger prion and amyloid diseases to switch from one stable conformation to another, with fatal outcomes.
To that end, a better understanding of the thermodynamics that guide the formation of macromolecular non-covalent bonds could help answer these questions, and also impact on various endeavors such as the rational design of protein-specific molecule drugs, self-organization of protein-inspired supramolecular structures, the composition of shape-memory biocompatible polymers and the development of stable protein therapeutics.
Differential scanning calorimetry (DSC) can be used to study and measure the thermodynamic parameters that control the formation of non-covalent bonds in proteins and other macromolecules. This article explores the application of DSC for the characterization of protein stability. Another calorimetric technique called isothermal titration calorimetry (ITC) is particularly useful for determining dynamic events such as kinetics and binding.
Analyzing Protein Thermal Stability
In order to study a protein’s stability in dilute solution, it is necessary to establish the changes that occur in the protein’s partial molar heat capacity at constant pressure (∆Cp). The change in heat capacity of a compound shows its ability to absorb heat and experience a defined rise in temperature. Water has an extensive network of hydrogen bonds and hence a higher heat capacity than organic compounds such as proteins. Water is also more organized and closely packed next to hydrophobic patches on the surface of the protein. As water is incapable of forming hydrogen bonds to nonpolar moieties, hydrogen bonding between the molecules of water is increased at the protein-solvent interface. As temperature increases, the organized water shell surrounding the protein starts to become disordered and becomes more like bulk water. This melting creates the anomalously large heat capacity of protein aqueous solutions. The protein’s contribution to the calorimetrically quantified heat capacity (its partial Cp) is calculated by deducting a scan of a buffer blank from the sample information before analysis. Besides being the heat capacity of protein, the partial Cp also includes any impact the protein had on the solvent, such as causing clathrate formation.
When the protein sample is heated, it first generates a slightly increasing baseline, but as heating continues, the protein absorbs the heat and thermally unfolds across a temperature range that is distinctive for that specific protein, creating an endothermic peak, as shown in Figure 1.
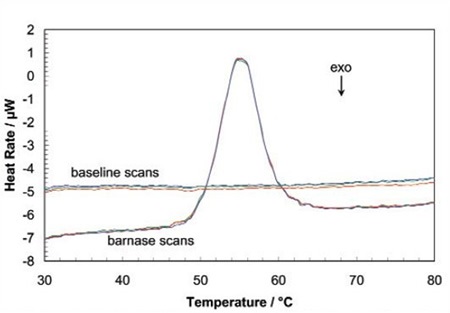
Figure 1. Three baseline temperature scans, and three scans of 60 μg of barnase in 0.3mL 20mM phosphate buffer, pH 5.5 obtained at a scan rate of 1 ˚C/min using a CSC6100 N-DSC II.
Throughout the unfolding process, molecules of water surrounding the protein reassemble and restructure as more non-polar side chains are exposed. Once unfolding is complete, the absorption of heat reduces and a new baseline is established. Following blank subtraction, the data can be studied to give a complete thermodynamic characterization of the unfolding event. Integration of the heat capacity of the sample versus temperature gives the enthalpy (∆H) of the unfolding event which is the result of exothermic events such as the disruption of hydrophobic interactions and endothermic processes such as the breaking of hydrogen bonds.
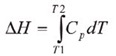
The shift in baseline before and after the transition represents the change in heat capacity (∆Cp) of the protein induced by unfolding. The transition midpoint (Tm,) refers to the temperature at which 50% of the protein molecules are folded and the rest are unfolded. The entropy (∆S) is obtained from the area under the curve of Cp/T vs.T.
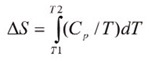
If only the determination of Tm is required, then it is not necessary to establish the reversibility of the unfolding reaction. However, to achieve a complete thermodynamic characterization of the unfolding event, the reversibility of the unfolding transition must be verified. This can be achieved by rescanning the sample under the same conditions and acquiring a second endotherm which can be superimposed on the first. If irreversibility is observed, it could be the result of a second unfolding process occurring following the first unfolding transition at Tm. The capillary cells in the CSC N-DSC II and N-DSC llI delay the onset of irreversible aggregation and precipitation of proteins, making it possible to obtain a complete and interpretable scan on any sample.
The sharpness of the transition peak reflects the cooperative nature of the unfolding process. If a narrow and relatively symmetric peak (Figure 2) is produced through the unfolding process, the transition will probably be reversible, two-state, and highly cooperative.
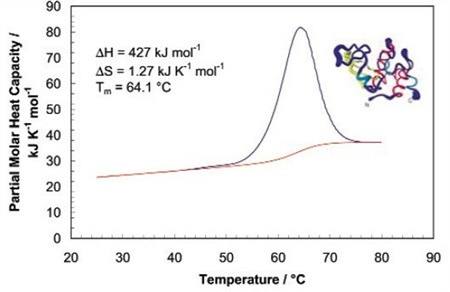
Figure 2. Temperature scan, converted to partial molar heat capacity, and fitted baseline for 1mg mL-1 hen egg white lysozyme in 0.2 M glycine buffer, pH 2.7. Data obtained at a scan rate of 1°C/min using a CSC 6100 N-DSC II.
This is often the case for small, one domain proteins. It is possible to verify two-state behavior by fitting the curve to the two-state van't Hoff equation. This calculation is carried out using software supplied with CSC DSCs. There can be a difference between the calorimetric enthalpy and the van't Hoff enthalpy obtained (∆HVH) because the former includes all contributions from the sample, including solvent rearrangement. If the calorimetric enthalpies and van't Hoff are the same, the denaturation process may be precisely approximated by the two-state model. If the van't Hoff enthalpy is larger than the calorimetric enthalpy, the protein is probably multimeric, and if it is smaller than the calorimetric enthalpy, an unfolding intermediate is probably formed.
Correlating Thermodynamic Properties to Stability
The protein’s stability and hence its Tm depend on environmental conditions. The protein’s conformational stability may be influenced by its concentration (demonstrating the significance of protein-protein interactions), scan rate (showing that thermal denaturation has a kinetic component) and ionic strength of the solution (which demonstrates the role of electrostatic interaction).
A full thermodynamic characterization of a lysozyme shows the complex interplay of stabilizing and destabilizing interactions. The different heat capacities of the folded and unfolded states show that the entropies and enthalpies of both forms are largely dependent on temperature, but they also compensate each other so that the net stability of a typical small protein is about 80 to 120kj/mol. Additionally, the large ∆Cp of unfolding shows that a protein will have a maximum stability at a given temperature (based on its pH, sequence etc.) and that its net stability will reduce above and below that given temperature. Therefore, both heating and cooling can denature proteins.
The quantity of protein available for characterization can be very restricted, which has previously limited the use of techniques such as DSC. However, new developments in instrument sensitivity have helped overcome this limitation by reducing the amount of sample needed for DSC studies.
Figure 3 illustrates the raw data for lysozyme scanned on a CSC N-DSC III instrument. The amounts of protein ranged from 400µg to 2µg in the 300µL sample cell. Thermodynamic parameters acquired at all concentrations are consistent and reliable (Table 1). This demonstrates that even 2µg can be enough to enable precise thermodynamic characterization of proteins.
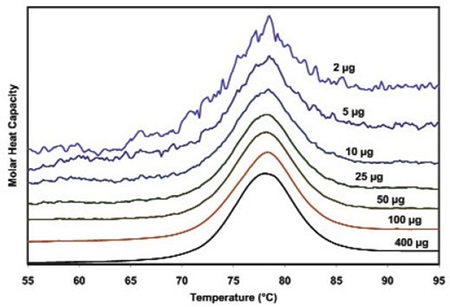
Figure 3. Temperature scan, converted to molar heat capacity, for hen egg white lysozyme solution in 0.2 M glycine buffer, pH 4.0. Lysozyme concentrations ranged from 400 to 2µg in the 300µL sample cell. Curves are offset to aid presentation. Data were obtained using a scan rate of 20C/min on a CSC 6300 N-DSC III.
With more proteins being characterized computationally and experimentally, correlations between structure and thermodynamics are being revealed, although not at a reliably predictive level as yet.
Rationalizing the Stabilities of Folded Conformations: Mutant vs. Wild-Type Proteins
Propensity scales for amino acids and hydrophobicity scales of the amino acids in many secondary structures are used along with molecular modeling to predict the impact of certain mutations. Systematic mutation of proteins that have known 3D structures often aims to quantify the ensuing stability changes in terms of the energetics of the unfolding process. Just one site-specific mutation can cause a significant decrease or increase in stability, but only small effects are produced by most mutations. Proteins are capable of making subtle conformational and solvation changes in order to compensate for the entropic and enthalpic disturbances induced by a mutation, which can mean only small free energy changes may be observed upon thermal protein denaturation. Nevertheless, the growing database of proteins characterized by DSC aids in understanding the effects mutations have on protein stability.
The latest redesign of proteins for improved stability and particularly the rational redesign of enzymes for improved entropic stability and catalytic function show that providing the mechanisms that control the stability and thermal unfolding of a protein are properly understood, the properties of proteins with a known structure can be logically and predictably changed.
Conclusion
It is important to understand the thermodynamics that control the formation of macromolecular noncovalent bonds in order to realize various endeavors such as the design of therapeutic proteins and supra-molecular assemblies. A number of factors contribute to protein stability, including hydrogen bonds to the solvent and intra-molecular interactions. Rigorous characterization of the thermodynamic basis of a protein’s stability is needed to improve its functional and thermal characteristics through sequence modification. DSC is the most direct approach for establishing protein stability in dilute solution, making it possible to quickly establish the relationship between entropy and enthalpy of the denaturation process. In addition, the superior sensitivity of the N-DSC III or CSC N-DSC II enables precise thermodynamic parameters to be obtained from just a few micrograms of protein.
Acknowledgement
Created from articles authored by Christin T. Choma, TA Instruments, 109 Lukens Drive, New Castle, DE 19720, USA.
References and Further Reading
(Preference has been given to current references. Citation does not imply that a paper is necessarily the original reference to a study.)
- Acevedo, O. E. and L. R. Lareo. (2005) Amino acid propensities revisited. OMICS 9,391-399.
- Anfinsen, C. B. (1973) Principles that govern the folding of protein chains. Science 181,223-230.
- Bruylants, G., J. Wouters and C. Michaux. (2005) Differential scanning calorimetry in life sciences: thermodynamics, stability, molecular recognition and applications in drug design. Curr. Med. Chem. 12,2011 -2020.
- Chan, H. S., S. Shimizu and H. Kaya. (2004) Cooperativity principles in protein folding. Methods Enzymol. 380, 350379.
- Cooper, A., M. A. Nutley and A. Wadood. (2001) Differential scanning calorimetry. p. 287-318. In S. E. Harding and B. Z. Chowdry (Eds.) Protein-Ligand Interactions: hydrodynamics and calorimetry. Oxford University Press, Oxford.
- Eijsink, V. G. H. et al. (2004) Rational engineering of enzyme stability. J.Biotechnol. 113,115-120.
- Fersht,A. R. and L.Serrano. (1993) Principles of protein stability derived from protein engineering experiments. Curr. Opin. Struct. Biol. 3,75-83.
- Fowler, S.B.ef al. (2005) Rational design of aggregation-resistant bioactive peptides: reengineering human calcitonin. PNAS 102,10105-10110.
- Freire, E. (1995) Thermal denaturation methods in the study of protein folding. Methods Enzyme/. 259,144-168.
- Griko, Y. V., E. Freire, G. Privalov, H. Van Dael and P. L. Privalov. (1995) The unfolding thermodynamics of c-type lysozymes: a calorimetric study of the heat denaturation of equine lysozyme.J. Mol. Biol. 252,447-459.
- Koehl, P. and M. Levitt. (1999) Structure-based conformational preferences of amino acids. PNAS 96,12524-12529.
- Langer, R. and D. A. Tirrell. (2004) Designing materials for biology and medicine. Nature 428,487-492.
- Lee, C-F., G. I. Makhatadze and K-B Wang. (2005) Effects of charge-to-ala nine substitutions on the stability of ribosomal protein L30e from Thermococcus celer. Biochemistry 44,16817-16825.
- Marky,L.A.andK.J.Breslauer.(1987) Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers 26,1601-1620.
- Mastrianni, J. A. (2004) Prion diseases. Clinical Neurosci. Res. 3, 469-480.
- Mendes,J., R.Gueroisand L.Serrano.(2002) Energy estimation in protein design. Curr. Opin. Struct. Biol. 12,441-446.
- O'Fagain, C. (2003) Enzyme stabilization: recent experimental progress. EnzymeMicrob.Technol. 33,137-149.
- Perl, D. and F. X. Schmid. (2002) Some like it hot: the molecular determinants of protein stability. ChemBiochem. 3,39-44.
- Pfeil,W.and P.L.Privalov.{1976) Thermodynamic investigations of proteins III. Thermodynamic description of lysozyme. Biophys. Chem. 4,41 -50.
- Plum, G. E. and K. J. Breslauer. (1995) Calorimetry of proteins and nucleic acids. Curr. Opin. Struct. Biol. 5,682-690.
- Privalov, P.Land N. N. Khechinashvili. (1974) A thermodynamic approach to the problem of stabilization of globular protein structures calorimetric study. J. Mol. Biol. 86,665684.
- Privalov,G.P.andP.L.Privalov.{2000) Problems and perspectives in microcalorimetry of biological macro molecules. Methods Enzymol. 323,31 -62.
- Remmele, R. Land W.R.Gombotz.(2000) Differential scanning calorimetry: A practical tool for elucidating stability of liquid pharmaceuticals. BioPharm. 13,36-46.
- Robertson, A. D. and K. P. Murphy. (1997) Protein structure and the energetics of protein stability. Chem. Rev. 97, 12511267.
- Russ, W. P. and R. Ranganathan. (2002) Knowledge-based potential functions in protein design. Curr. Opin. Struct. Biol. 12,447-452.
- Scharnagl,C.,M.Reif and J. Friedrich. (2005) Stability of proteins: temperature, pressure and the role of solvent. Biochim. Biophys.Acta.1749,187-213.
- Sela, M., F. W. White and C. B. Anfinsen. (1957) Reductive cleavage of disulfide bridges in ribonuclease. Science 125, 691-692.
- Shinoda, K. (1977) 'Iceberg' formation and solubility. J. Phys. Chem. 81,8069-8072.
- Sturtevant, J. M. (1994) The thermodynamic effects of protein mutations. Curr. Opin. Struct. Biol. 4,69-78.
- Yeates,T. O.and J. E. Padilla. (2002) Designing supramolecular protein assemblies.Curr. Opin.Struct. Biol. 12,464-470.
- Webeter, D. M. (Ed.) (2000) Protein structure prediction. Humana Press.
About TA Instruments
 TA Instruments' reputation for high technology products, quality manufacturing and unbeatable after sales support is why more customers recommend TA products to their colleagues around the world. Headquartered in New Castle, DE, TA Instruments prides ourselves in the technical competence and professionalism that our sales force offers. We are the only thermal analysis, rheology, and microcalorimetry supplier recognized worldwide for our prompt, courteous and knowledgeable service staff, the hallmark of our company. Our technical support group is committed to handling all of your thermal analysis and rheology needs, and is available by phone, email and through the internet.
TA Instruments' reputation for high technology products, quality manufacturing and unbeatable after sales support is why more customers recommend TA products to their colleagues around the world. Headquartered in New Castle, DE, TA Instruments prides ourselves in the technical competence and professionalism that our sales force offers. We are the only thermal analysis, rheology, and microcalorimetry supplier recognized worldwide for our prompt, courteous and knowledgeable service staff, the hallmark of our company. Our technical support group is committed to handling all of your thermal analysis and rheology needs, and is available by phone, email and through the internet.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.