Sponsored Content by Bruker BioAFMReviewed by Mychealla RiceDec 22 2022
The primary task of T lymphocytes during an immune reaction is to inspect the surface of neighboring cells and identify the presence of external peptide pathogens on antigen-presenting cells (APC) through the membrane T cell receptor (TCR) among the numerous self-peptides introduced by the Major Histocompatibility Complexes (MHC).
A TCR-peptide-MHC (pMHC) contact is essential for stimulating T-cells and associated activities, such as proliferation, at the heart of the adaptive immune system response.
TCR-pMHC interactions also provide continual survival signals to keep a consistent population of memory cells, which comprise the long-term immunity system.1
TCR-dependent signaling must be quick and accurate for detecting the presence of relatively few foreign peptide antigens while filtering out self-peptide/MHC-generated noise, so as not to damage healthy cells and tissue.
Image Credit: Bruker BioAFM
For the formation and operation of the adaptive immune response, the issue of how TCR-peptide-MHC binding events are interpreted by the lymphocytes' cellular signaling apparatus remains crucially relevant.
Significant work has been carried out to quantify the dynamics of the TCR-peptide-MHC bonds, the number of interacting proteins, the type, and contribution of co-stimulatory particles, the spatial organization of the stimulating particles, and the involvement of other physical variables, such as the forces applied on the molecular bonds.2
The cytoskeletal architecture of T cells is essential in its identification and activation capabilities, although it must be further clarified and quantified. It permits the T cells to apply forces on the APC at an individual molecule level.
These forces are crucial in developing the ability of T cells to identify foreign peptides selectively and sensitively and to activate them. In addition to the substrate's physicochemical features, T cells employ the substrate's general stiffness as an additional signal.
Mechano-transduction is believed to function in the activation of T cells, at both molecular and cellular scales.3
The purpose of these studies is to analyze how the micro- and nano-organization of activation-related surface particles influence or are influenced by the micromechanical characteristics of T cells (such as identification, elasticity, and membrane tension) by employing modern biophysical methods based on force application such as atomic force microscopy (AFM) or optical tweezers (OT).
During signaling, these approaches allow the evaluation of cell elasticity, viscosity, and adherence. OT and nano-indentation by AFM enable the researcher to quantify the elasticity and viscoelasticity of cells. Membrane tension can also be detected by using AFM or OT to pull tethers from the membrane.
Both AFM and OT are essential tools in the area of immuno-mechanics, as will be shown in the next sections.
Evaluation of TCR recognition at the single molecule scale on living T cells
To establish which physical properties are recognized by T cells when they come in contact with an antigen-presenting cell, a single molecule investigation was designed to identify the likelihood of adhesion and the forces of separation at the surface of living lymphocytes.
Single-molecule force spectroscopy (SMFS) was used in this study and the tips of soft AFM cantilevers were embellished with duplicate pMHC molecules (Fig. 1).
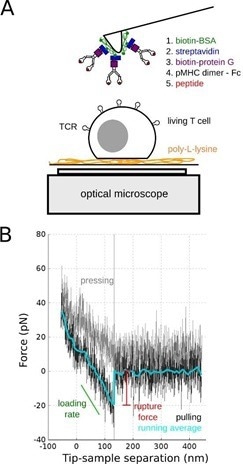
Figure 1. SMFS of TCR/pMHC bond. A: Schematics of the experiments on living T cell hybridoma, with the methodology used to decorate AFM cantilevers with pMHC. B: Typical single molecule separation event, showing the very low specific forces (~ 20 pN) that were recorded, to be compared to larger forces obtained for classical adhesion molecules. Adapted from [4]. Image Credit: Bruker BioAFM
These were used to investigate the mechanics of the T cell under extremely low stresses and for brief contact times (~ 100 msec). As a consequence, recognition events were rare.
By changing the peptide load in the MHC pocket, it was demonstrated that the recognition was specific. However, the rupture forces seen under the experimental settings were independent of the peptide.4
Using methods with softer springs (a laser for OT; a red blood cell for a biomembrane force probe), it was subsequently demonstrated that the TCR might act as a mechanosensory protein. It is currently believed that the forces felt by the T cell through the TCR contribute to its extremely specific and quick identification.5
Real-time combination of AFM and fluorescence microscopy
AFM indentation of T cells was concurrently performed with intracellular Ca2+ fluorescence imaging to quantify the forces at play throughout the different activation stages. 6
A unique internal timer signal and AFM were used to apply a mechanical stimulation that can be made using anti-CD3 antibodies.
Micro-manipulation techniques were developed for decorating AFM cantilevers with beads ranging in diameters from 1 to 50 µm to regulate the shape and size of the stimulating interface.
To investigate how the “signaling black box” responds to the same precise signal, but with a predetermined spatial or temporal distribution, stimulations can be conducted with either constant contact or a succession of timed, short stimulations.
This approach is a conventional physicist technique for analyzing the “transfer function” of an unknown system. Here, biology enables the tampering of the “black box” by using mutants or chemicals that can affect specific parts of the signaling cascades.
Using optogenetic techniques, such as photoactivatable Rac, a small GTP-binding protein, protocols were developed to record the alteration of the mechanical properties of T cells.6
Characterization of substrates for interaction with T cells
Two AFM modes may be used to analyze the topological or mechanical characteristics of a synthetic substrate that will interact with the T cells. Initially, one can image the substrate at the nanometer scale and examine its roughness or surface structure, such as by micro-contact printing or nano-scale patterns.7,8
Secondly, when developing soft polymeric substrates or soft gels, substrate indentation or force mapping may be employed to accurately characterize the mechanical characteristics of the substrate the cell is exposed to using a thin tip/small scan range at the subcellular scale or using a bead for cellular contact/larger scan range at the cellular scale.
The mechanical characteristics of the substrate can cause T cells to spread unusually based on the molecular decoration present at the surface.3
Characterization of model antigen-presenting cells
A model APC (COS-7 cells) was formed on stamped fibronectin while limiting the attachment and activation of the T cells.9 Micropatterns were utilized to tug on the cell’s edge, resulting in a large lamellipodium thin enough to allow sophisticated surface microscopies such as total internal reflection fluorescence (with membrane-labeled T cells) or reflection interference contrast microscopy (RICM) (without any labeling) for observing the early interactions of T cells.
This structure presents a completely regulated, dynamic, cellular environment. The composition of this environment is adjustable by transfection and is closer to physiological settings than textured substrates and supported lipid bilayer techniques.10
The lamellipodium’s 3D topography was studied using different patterns and a mixture of TIRF, confocal (membrane, cytoskeleton), and multi-color RICM reconstruction, in combination with AFM scanning and force measurements.11
Traditional AFM indentations were used to quantify Young’s modulus of diverse populations of COS cells expressing various receptors and molecules in terms of T cell mechanics (unpublished data, coherent with reference 12).
T lymphocyte/Antigen presenting cell forces
To more closely imitate physiological conditions, the APC vs. T cell issue was addressed by connecting a model APC (a transfected COS-7 cell) to an AFM cantilever and bringing it in contact with a (Ca2+ reporter loaded) T cell of known shape.9
This approach, known as single-cell force spectroscopy (SCFS), allows for the assessment of the contact mechanics between cells, the development of the contact, and the forces required for separation, at sizes ranging from single molecules to full cell scales.14
It has recently been used in T cell/APC interaction during long contact times and after the development of the immune synapse.13 However, adhesion was only recorded at the cellular scale, and simultaneous observation of the early stimulation of the cells was not carried out.
Integrating SCFS with the as-described technique of concurrent AFM/fluorescence for recording the activation pattern before, during, and after contact, can enable the investigation of the initial physical determinants of T cell activation, such as which forces are needed or developed by T cells for integrating a biochemical or biomechanical signal (Fig. 2).
Such techniques may need a long piezo range (up to 100 µm or more) based on the contact periods, molecules involved, and possible signals required to strengthen the cell/cell adhesion.
It may also be essential to synchronize the fluorescence-detecting lens with a supplementary piezo. All of this is possible with the JPK CellHesion module, an add-on of the JPK NanoWizard AFM.14
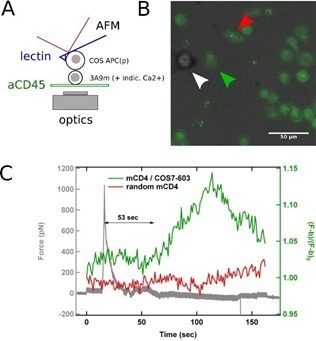
Figure 2. SCFS coupled to fluorescence microscopy. A: Schematics of the experiments, where a calcium reporter loaded T cell, gently immobilized on a PLL-coated surface, is brought into contact with an APC attached to an AFM lever. B: Superposition of transmission and fluorescence images, with the APC (white arrowhead), the contacted T cell (green arrowhead) and a control/non-contacted T cell (red arrowhead). C: Force signal, presented with the fluorescence signal emitted by contacted and control T cells. The rise in calcium follows the contact, as detected by force rise, by ~ 1 min, which is coherent with literature. Image Credit: Bruker BioAFM
Cell/cell adhesion forces, numbers, and detachment distances are greater than in conventional SCFS (cell vs. substrate) experiments, necessitating the adjustment of the cantilever’s spring constant and the puling range, which is generally restricted to 10-15 µm in traditional AFMs.
Measuring T cell cortical tension by pulling membrane tethers
AFM can pull membrane tubes or tethers by using lectin-coated cantilevers. Cortical cell tension can be measured through the forces required to remove them from the cell.
Using drug-induced disturbance of the cell cytoskeleton in a parallel investigation, the cell’s membrane tension and the binding energies between the membrane and the cytoskeleton can also be assessed.15,16
The resolution in force can be compared to an AFM using optical tweezers (the JPK NanoTracker 2). These investigations can be undertaken with fluorescence imaging of the membrane (Fig. 3). Detailed measurements of the tube pulling mechanics are possible due to the very low noise level (~ pN).
A lengthy pulling distance may be required if T cells are small (~10 µm in diameter). However, tethers up to 10 or even 20 µm long may be pulled depending on the experimental settings.
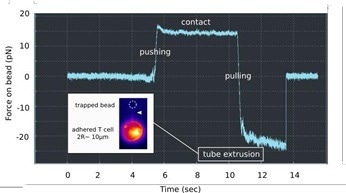
Figure 3. Membrane tube pulling using OT. A lectin-coated bead is used to contact a PLL immobilized T cell, and to pull a long membrane tether, as denoted by a force plateau in the retraction (« pulling ») part of the force curve vs. Time. Insert: Fluorescence monitoring of the membrane tube (white arrowhead), using a mutant cell line, the membrane of which is fluorescent. Image Credit: Bruker BioAFM
Conclusions and perspectives
T cells exhibit mechanosensory characteristics, ranging from the TCR molecule to the entire cell size.2
Several examples have been presented demonstrating how AFM, in imaging or force mode, is a very useful tool for characterizing the surface with which the T cells come into contact, as well as mechanics and forces of the T cells themselves.
AFM may also be used in conjunction with fluorescence microscopy for real-time analysis of the intracellular signals created when the proteins on the cell surface are activated.
Completed characterization of cellular dynamics (Young’s modulus, tension, and by using oscillating mechanical modes G’ and G’’ - dynamic shear modulus) can be acquired by combining AFM with OT.
Information on the sub-membrane structure of the cytoskeleton can also be obtained by altering the size and form of the indentor at multiple scales. Interesting possibilities arise when the systems are combined, including:
(i) Real-time fluorescence, AFM, and OT can lead to the analysis of the membrane/surface protein of the system’s transfer function.6 When AFM imaging is combined with fluorescence microscopy, 3D functional structures on the cell surface can also be studied.14
(ii) Combining AFM with OT in a unified system enables the reproduction of accurate physiological environments, as one can assess the contact between a T cell and an antigen-presenting cell using SCFS. The modulation of the recognition forces can be recorded when a secondary T cell is brought into contact.17
Overall, the use of nano-force instruments holds great potential for the burgeoning disciplines of immuno-biophysics and immuno-mechanics.
Authors
Produced from materials originally authored by Anais Sadoun1, Farah Mustapha1, Omar Ndao1, Gautier Eich1, Yannick Hamon2, Laurent Limozin1, and Pierre-Henri Puech.1
1 Aix Marseille University, LAI UM 61, Inserm, UMR_S 1067, CNRS, UMR 7333, Turing Center for Living Systems, Marseille, F-13288, France.
2 Aix Marseille Univ, CNRS, INSERM, CIML, Marseille, France
Acknowledgments
This work was supported by funding from Prise de Risques CNRS (PHP), the ANR JCJC DissecTion (PHP), and from the Labex INFORM (ANR-11-LABX-0054, to Inserm U1067 Lab & CIML, and as Ph.D. grants to AS & FM). This project has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No713750. Also, it has been carried out with the financial support of the Regional Council of Provence- Alpes-Côte d’Azur and with the financial support of the A*MIDEX (n° ANR- 11-IDEX-0001-02), funded by the Investissements d'Avenir project funded by the French Government, managed by the French National Research Agency (ANR) (as a Ph.D. grant to FM). It was also supported by GDR Imabio as grants to ON & GE.
References and Further Reading
- Murphy, Ken, Paul Travers, and Mark Walport. 2008. Janeway’s Immunobiology 7th Edition. Edited by Garland Science.
- Limozin, Laurent, and Pierre-Henri Puech. 2019. ‘Membrane Organization and Physical Regulation of Lymphocyte Antigen Receptors: A Biophysicist’s Perspective’. The Journal of Membrane Biology 252 (4–5): 397–412. https://doi.org/10.1007/s00232-019-00085-2
- Wahl, Astrid, Céline Dinet, Pierre Dillard, Aya Nassereddine, Pierre-Henri Puech, Laurent Limozin, and Kheya Sengupta. 2019. ‘Biphasic Mechanosensitivity of T Cell ReceptorMediated Spreading of Lymphocytes’. Proceedings of the National Academy of Sciences of the United States of America 116 (13): 5908–13. https://doi.org/10.1073/pnas.1811516116
- Puech, Pierre-Henri, Damien Nevoltris, Philippe Robert, Laurent Limozin, Claude Boyer, and Pierre Bongrand. 2011. ‘Force Measurements of TCR/PMHC Recognition at T Cell Surface’. Edited by Daniel J. Muller. PLoS ONE 6 (7): e22344. https://doi.org/10.1371/journal.pone.0022344
- Huse, Morgan. 2017. ‘Mechanical Forces in the Immune System’. Nature Reviews Immunology 17 (11): 679–90. https://doi.org/10.1038/nri.2017.74
- Cazaux, Séverine, Anaïs Sadoun, Martine Biarnes-Pelicot, Manuel Martinez, Sameh Obeid, Pierre Bongrand, Laurent Limozin, and Pierre-Henri Puech. 2016. ‘Synchronizing Atomic Force Microscopy Force Mode and Fluorescence Microscopy in Real Time for Immune Cell Stimulation and Activation Studies’. Ultramicroscopy 160 (January): 168–81. https://doi.org/10.1016/j.ultramic.2015.10.014
- Lo Schiavo, Valentina, Philippe Robert, Zohar Mishal, PierreHenri Puech, Francesco Gentile, Paolo Decuzzi, Pierre Bongrand, and Laurent Limozin. 2013. ‘Transient Adhesion Mediated by Ligand-Receptor Interaction on Surfaces of Variable Nanotopography’. International Journal of Nanotechnology 10 (5–7): 404–18. https://doi.org/10.1504/IJNT.2013.053512
- Pi, Fuwei, Pierre Dillard, Laurent Limozin, Anne Charrier, and Kheya Sengupta. 2013. ‘Nanometric Protein-Patch Arrays on Glass and Polydimethylsiloxane for Cell Adhesion Studies.’ Nano Letters, June. https://doi.org/10.1021/nl401696m
- Salles A, Billaudeau C, Sergé A, Bernard A-M, Phélipot M-C, Bertaux N, et al. 2013. ‘Barcoding T Cell Calcium Response Diversity with Methods for Automated and Accurate Analysis of Cell Signals (MAAACS)’. PLoS Comput Biol 9(9): e1003245. https://doi.org/10.1371/journal.pcbi.1003245
- Dillard, Pierre, Rajat Varma, Kheya Sengupta, and Laurent Limozin. 2014. ‘Ligand-Mediated Friction Determines Morphodynamics of Spreading T Cells’. Biophysical Journal 107 (11): 2629–38. https://doi.org/10.1016/j.bpj.2014.10.044
- Dejardin, Marie-Julie, Arnaud Hemmerle, Anaïs Sadoun, Yannick Hamon, Pierre-Henri Puech, Kheya Sengupta, and Laurent Limozin. 2018. ‘Lamellipod Reconstruction by ThreeDimensional Reflection Interference Contrast Nanoscopy (3DRICN)’. Nano Letters, September. https://doi.org/10.1021/acs.nanolett.8b03134
- Bufi, Nathalie, Michael Saitakis, Stéphanie Dogniaux, Oscar Buschinger, Armelle Bohineust, Alain Richert, Mathieu Maurin, Claire Hivroz, and Atef Asnacios. 2015. ‘Human Primary Immune Cells Exhibit Distinct Mechanical Properties That Are Modified by Inflammation’. Biophysical Journal 108 (9): 2181– 90. https://doi.org/10.1016/j.bpj.2015.03.047
- Hosseini, Babak H., Ilia Louban, Dominik Djandji, Guido H. Wabnitz, Janosch Deeg, Nadja Bulbuc, Yvonne Samstag, Matthias Gunzer, Joachim P. Spatz, and Günter J. Hämmerling. 2009. ‘Immune Synapse Formation Determines Interaction Forces between T Cells and Antigen-Presenting Cells Measured by Atomic Force Microscopy’. Proceedings of the National Academy of Sciences of the United States of America 106 (42): 17852–57. https://doi.org/10.1073/pnas.0905384106
- Puech, Pierre-Henri, Kate Poole, Detlef Knebel, and Daniel J Muller. 2006. ‘A New Technical Approach to Quantify Cell-Cell Adhesion Forces by AFM.’ Ultramicroscopy 106 (8–9): 637– 644. https://doi.org/10.1016/j.ultramic.2005.08.003
- Diz-Muñoz, Alba, Michael Krieg, Martin Bergert, Itziar Ibarlucea-Benitez, Daniel J. Muller, Ewa Paluch, and CarlPhilipp Heisenberg. 2010. ‘Control of Directed Cell Migration In Vivo by Membrane-to-Cortex Attachment’. Edited by William A. Harris. PLoS Biology 8 (11): e1000544. https://doi.org/10.1371/journal.pbio.1000544
- Sadoun, Anaïs, and Pierre-Henri Puech. 2017. ‘Quantifying CD95/Cl-CD95L Implications in Cell Mechanics and Membrane Tension by Atomic Force Microscopy Based Force Measurements’. Methods in Molecular Biology (Clifton, N.J.) 1557: 139–51. https://doi.org/10.1007/978-1-4939-6780-3_14
- Chen, Jiahuan, Anutosh Ganguly, Ashley D Mucsi, Junchen Meng, Jiacong Yan, Pascal Detampel, Fay Munro, et al. 2017. ‘Strong Adhesion by Regulatory T Cells Induces Dendritic Cell Cytoskeletal Polarization and Contact-Dependent Lethargy’. Journal of Experimental Medicine, 1–12. https://doi.org/10.1084/jem.20160620
About Bruker BioAFM
Bruker BioAFM, former JPK Instruments AG, is a leading manufacturer of nano-analytical instruments - particularly based on atomic force microscope (AFM) and optical tweezers systems - for life sciences and soft matter applications.
We combine the highest technical skills with visionary applications. Our work applies nanotechnology in ways to provide solutions to challenges facing researchers in life sciences and soft matter today. Driven by inspiration and ambition, it is our conviction that only the best tools are good enough for the research of life. We are listening with the ear of a scientist in detail to the current challenges of our customers and find individual solutions for individual problems. This is how we understand our business.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.