Please note: This product is for research use only.
Lipoproteins can be described as supramolecular assemblies that carry water-insoluble lipids in the blood. They also carry apolipoproteins that determine both function and structure.
Apolipoproteins are a monolayer of cholesterol and amphiphilic phospholipids that are embedded within. They represent the surface of the lipoproteins that “hide” the lipids from the aqueous environment. The inner core of apolipoproteins mostly contains esterified cholesterol and triglycerides.
Lipoproteins are usually categorized into five major groups. These include:
- High-density lipoproteins (HDL)
- Low-density lipoproteins (LDL)
- Intermediate density lipoproteins (IDL)
- Very low-density lipoproteins (VLDL)
- Chylomicrons
With respect to the assessment of cardiovascular risks, the lipoprotein class-specific concentrations of triglycerides and cholesterol hold considerable interest beyond their overall concentrations inside the plasma. It has been strongly indicated that the prediction of cardiovascular risk can be improved by additional sub-classification of lipoproteins.
The preferred method is based on the investigation of signals in the proton NMR spectrum associated with the lipoproteins. Variations in the size, composition, and density of lipoproteins translate into respective differences in signal line shapes. Such differences can be used to obtain data on both main and subclasses of lipoproteins (refer to Table 1).
It is essential to develop a regression model using a training data set. This data set contains:
- Proton NMR spectra of the same set of samples
- Lipoprotein analytes from total plasma as well as main and subclasses based on ultracentrifugation
After the regression model is determined, the lipoprotein analytes from the proton NMR spectra of serum samples or new plasma are directly calculated by a prediction algorithm, without any need for ultracentrifugation.
Through this proton NMR method, data about lipoprotein-related information can be obtained on:
- The main HDL, LDL, IDL, and VLDL classes
- Four HDL-subclasses such as HDL-1 to HDL-4
- Six LDL sub-classes such as LDL-1 to LDL-6
- Six VLDL subclasses such as VLDL-1 to VLDL-6 (respectively sorted in accordance with increasing density and decreasing size)
- Serum and plasma
Data includes concentrations of lipids, that is, free cholesterol, cholesterol, triglycerides, and phospholipids; concentrations of apolipoproteins Apo-B, Apo-A1, and ApoA2; and the LDL particle numbers. All parameters determined by the proton NMR lipoprotein analysis are shown in Table 1. The front page of the automatic report (B.I.-LISA) is shown in Figure 1.
Table 1. Listing of all parameters generated by the proton NMR lipoprotein subclass analysis. Source: Bruker Biospin Group
Figure 1. Page 1 of a B.I.-LISA report covering the main fraction information. Image Credit: Bruker BioSpin Group
Validation of the Lipoprotein Subclass Analysis
The following measures were considered and constantly followed to test the validity of the results:
- Laboratory comparison tests
- Test transferability of measurement and analysis
- Traceability tests on key parameters utilizing certified reference samples
- Test reproducibility of measurement and analysis (refer to Figure 2)
Image Credit: Bruker BioSpin Group
Advantages of Bruker B.I.-LISA IVDr Lipoprotein Analysis
- Fully automated and easy to run (for example, by an MTA)
- Relatively faster turnaround time - minutes against days
- Equal specificity and sensitivity as that of ultracentrifugation
- Costs less than ultracentrifugation method
- Simple sample preparation
- A small volume of samples (500 µL) - improved patient compliance
- Spectra can be utilized for different types of analysis
- There is no direct contact between the device and the sample
Lipoprotein Subclass Analysis with NMR
The present testing modality, for example, the ultracentrifugation method, usually takes one week for each sample. By contrast, the Bruker B.I.-LISA lipoprotein panel provides considerable benefits. Currently, the ultracentrifugation method can manage up to 16 samples in parallel, but the process demands accurate and repeated manual interaction. With respect to VLDL subclass analysis, an ultracentrifuge with modifiable angles is needed. Therefore, alternative methods are usually sought.
Bruker has headed a study to find out lipoprotein subclass analysis based on proton NMR spectroscopy of serum samples or blood plasma, utilizing regression analysis of ultracentrifugation results.
The aim was to offer an analytical technique that:
- Offers adequate precision at maximum reproducibility and provides complete transferability from one instrument to another
- Operates at high throughput (up to 150 samples each day in high-quality regular screening) at a lower cost for each sample, under simple sample preparation, complete automation, and automatic report generation
- Determines LDL particle numbers, apolipoproteins A1, A2, B, triglycerides, phospholipids, and cholesterol for both main and subclasses of lipoproteins for serum and plasma.
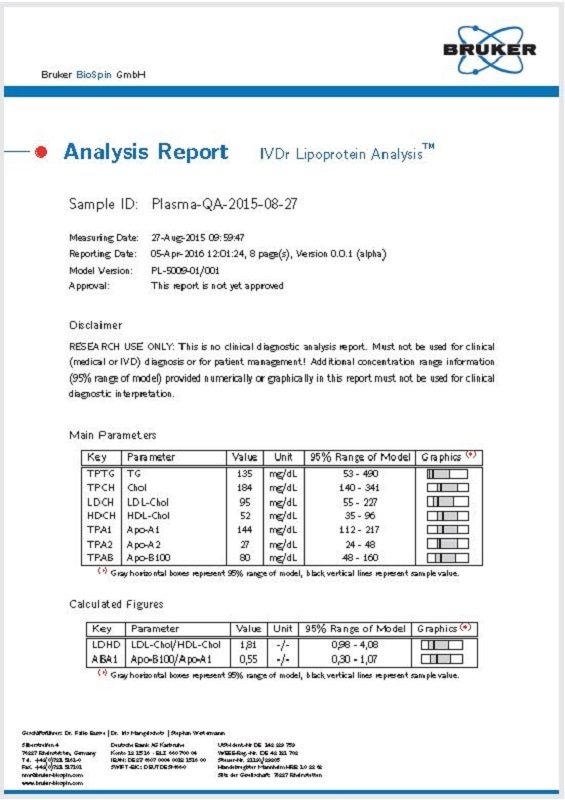
Figure 2. Short and long-term imprecision of lipoprotein measurements by NMR (NCEP National Cholesterol Education Program, United States). Image Credit: Bruker BioSpin Group
It must be noted that the analytes shown in Table 1 are partly correlated, and therefore cannot be taken as independent analytes. The association between the analytes relies on only a few latent factors. These factors are established by lipid metabolism as well as the limits of the training data set from the ultracentrifugation method utilized for training the regression model. This regression model has been exclusively created for 600 MHz spectra.
Use of Lipoprotein Subclass Analysis in Translational and Clinical Research Applications
- Stroke
- Thrombosis
- Atherosclerosis
- Fatty liver disease
- Obesity, Type 2 diabetes, and metabolic syndrome
- Cardiovascular diseases (studies on early detection, grading, prevention, and treatment)
- Cancer
- Inflammatory diseases
- Cerebrovascular diseases
- Epidemiological studies
- Effect of food on health
- Quality control of serum/plasma, fasting state, biobanks, and concentration panel with spectra
Figure 3. Two examples of lipid distributions calculated from Bruker IVDr Lipoprotein Subclass Analysis B.I.-LISA. Image Credit: Bruker BioSpin Group
Requirements for Lipoprotein Subclass Analysis
- Use of Bruker SOPs for serum/plasma
- IVDr platform at 600 MHz
- Access to Bruker Data Analysis server for completely automated remote analysis (spectra transfer following measurement to Bruker server through private FTP, back-transfer of result report)
- Solvent suppression, absolute temperature, and quantification of the reference sample should be checked routinely (preferably every day)