Can you give a brief overview of intrinsically disordered proteins (IDPs)? In your opinion why are IDPs so significant?
“IDPs” is now a widely used acronym that stands for “intrinsically disordered proteins.” It is the term generally used by the scientific community to refer to a wide variety of proteins that do not have a stable 3D structure and are instead characterized by a high extent of local mobility, disorder and many conformers that are accessible at room temperature.
These are all very peculiar features that confer them a variety of functional advantages in respect to those derived from the presence of well-defined 3D structures.
Focusing on IDPs at CERM
Focusing on IDPs at CERM from AZoNetwork on Vimeo.
Why is research into IDPs so far behind the research into structured proteins?
Until about 15 years ago, these proteins were not much considered, although they have always existed. An example is casein, which we all drink in the morning when we have our cappuccino, as it is present in milk.
Tools that have been used for 50 years to determine 3D structures such as X-ray and NMR, attracted attention more towards folded proteins.
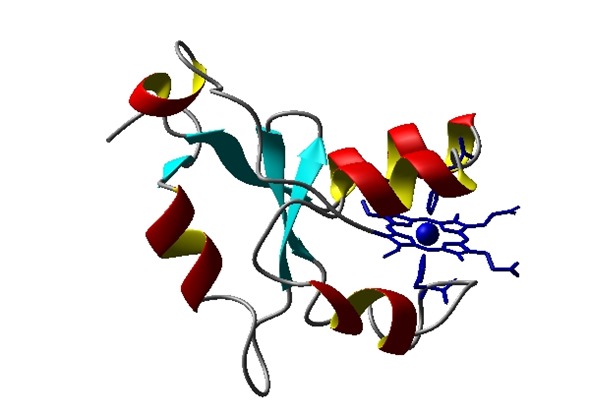
Structural model of folded protein structure of cytochrome b5
What differences in structure and function do IDPs have over non-intrinsically disordered proteins?
We all grew up reading in textbooks about how the function of a protein is primarily linked to its 3D structure. A lot of data produced since the 1950s about the 3D structures of proteins has been deposited in the protein databank (PDB) and explains a lot of features about the function of the proteins themselves.
This mainstream thinking did distract the scientific community from also looking into other kinds of proteins, proteins that do not have a well-defined 3D structure in their native form, but that inter-convert between a variety of different conformations, with backbones that are largely solvent exposed, highly flexible and highly disordered.
It seems very simple to say it now, but it's pretty obvious that these very different properties confer completely different functional advantages.
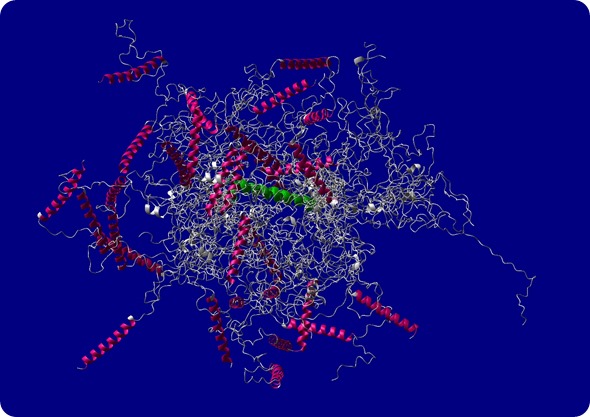
Structural model of intrinsically disordered protein ID4
How was the difference between these two classes of proteins treated within the scientific community? How have the latest imaging techniques shaped the way that these proteins have been studied?
One thing I enjoyed reading about in a review by our colleague Vladimir Uversky, one of the people who opened this field, was about some of the names that have been used since the 1950s to refer to these proteins that were not really the main focus of the scientific community.
He found a variety of different names including “malleable,” “pliable” and different combinations of the terms “disordered,” “unfolded” with “intrinsically,” native”. Finally, some more creative terms were used such as “dancing proteins,” “protein clouds” and “proteins waiting for partners.”
It was good that about ten years ago, the scientific community agreed on a general term that would somehow broadly indicate this class of proteins that do not fold in a well-defined and stable 3D structure, like we were used to thinking before.
While focusing on this subject of IDPs, I was really impressed by how all we know is largely driven by the technology that we have and that we can exploit. I think this investigation of proteins at atomic resolution gives a clear example.
We have been able to do X-rays of crystals since, essentially, the 50s but, on the other hand, we didn't have a tool to really measure dynamics at atomic resolution. Thanks to X-ray and NMR determining 3D structure in a fairly easy manner, the community was pushed to focus more and more on the study of folded proteins.
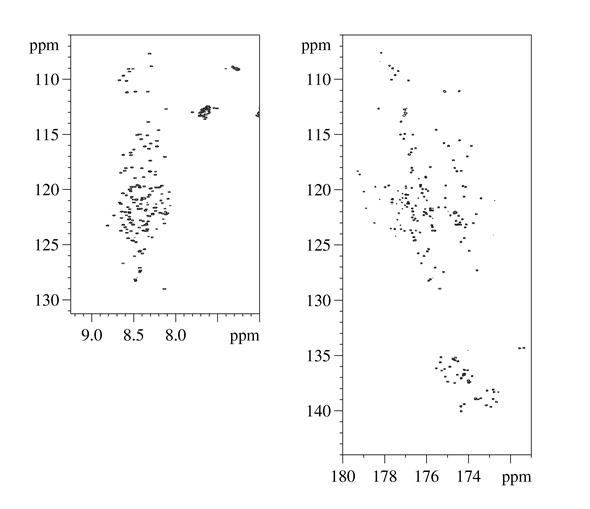
2D HN and 2D CON NM spectra acquired for intrinsically disordered domains of CBP (CBP_ID4)
Over the years, data has accumulated in the protein data bank that explains a variety of different functions and has contributed a lot to improving our knowledge about the properties of folded proteins.
Why does NMR hold a strategic role in IDP characterization? What advantages are there that NMR delivers that other instrumentation can’t?
This mainstream understanding kept the scientific community distracted from also focusing on a lot of other important proteins, such as the more flexible ones. We all know how NMR spectroscopy is not only used to look at structural information, but can also provide a variety of information about local dynamics and flexibility, so it can now play such a strategic role in the study of IDPs.
It can contribute to enabling access to high resolution information on these proteins and help to fill a gap of about 50 years in terms of what we know about these proteins.
On the other hand, to focus on intrinsically disordered proteins, you need a high resolution because the properties of the proteins such as high flexibility, provide resonances that are all very close to one another.
This typically leads to the problem of extensive cross-peak overlaps in the spectra. Therefore, high fields, isotopic labeling and tailored experiments are very important to improve the methods to study IDPs.
We all know that NMR is great for providing both structural and dynamic information and that's why it's a strategic technique in general to study highly flexible and dynamic systems, in particular IDPs, which are also quite complex. Despite this, I think we can do a lot better if we think about how these properties impact on NMR parameters.
They will show us some critical points that we have not been thinking too much about due to focusing more on the study of folded proteins. If we think carefully about the impact of the properties of IDPs such as the highly flexible backbones, the largely exposed amide protons to the solvent and their high dynamicity, then we can do a lot to further improve the NMR experiments so that we can study IDPs of increasing complexity.
Can you give an introduction into your recent research using NMR to study IDPs? What applications are there of this research?
Our interest in IDPs came from looking at the properties of NMR spins. We have been focusing on the development of new methods based on carbon 13 direct detection. This is thanks to a strong interaction with Bruker since, in order to focus on carbon detection, the sensitivity of the instrumentation is really important.
When we started to focus on the project, we tried to see which machine in the lab had the highest carbon sensitivity. We were really surprised that the older the probe, the better the carbon sensitivity.
Although surprising at the time, it actually made sense because, over the years, the probes have been largely improved for proton sensitivity rather than carbon sensitivity. We therefore used the oldest probes for our first experiments.
Then, working with a close interaction with the people at Bruker, with both application specialists and those working on the construction of the probe, they asked us what should be improved.
We suggested new applications and over a very exciting 10 years, step by step, we managed to go from a sensitivity of 200 to 1 with carbon 13, to the sensitivity we have now, which is 2800 to 1.
This means an improvement of a factor of 14 over a period of less than 10 years. That definitely enables a huge amount of applications based on this technology. Just to give an idea in terms of the amount of time you need to apply a specific experiment, if the sensitivity improves by a factor of 10, the amount of time you need is reduced by a factor of 100.
This means that now, we can really use these experiments based on carbon detection, which we've been focusing on over the last 10 years, as a routine tool to study proteins in general. That is something that is particularly useful for proteins where you need some complimentary tool for proton detection experiments.
Motivated by the need for new methods to focus on paramagnetic proteins, we started to focus on this subject. That was traditionally the focus of CERM. It was started by Ivano Bertini.
Ivano, Claudio and Lucia have vast experience in dealing with paramagnetic proteins, as do many of my colleagues here at CERM like Paola, Mario and Roberta. Initially, we did a lot of work with paramagnetic proteins, but then, when studying the properties of the spins, we realized that these methods could be interesting, not only for detecting signals near the paramagnetic center of proteins, but also, for looking at very large systems or for studying IDPs, for example. This is how we became interested in the study of IDPs.
Over there on the board, is a CON spectrum of alpha synuclein, which is well known and well characterized and has now become the sort of standard sample for the setup of experiments on IDPs. It's very interesting because it's involved in the progression of neurodegenerative diseases. It has been used for methods development in our lab.
Over the last 10 years, at first we focused on our major topic of carbon direct detection, but then we approached the field of IDPs, looking at all their peculiar properties, particularly the high flexibility, the largely solvent exposed backbones and the high dynamicity.
We wanted to know how these properties impact on NMR observables and why, because, if you know that, then you can design better experiments to broaden the range of applications of NMR to the study of IDPs.
What were the aspects that we found important within the study of IDPs?
One well known problem is high cross-peak overlap. We explored heteronuclei (13C and 15N) as much as possible, since these are characterized by a higher chemical shift dispersion.
This was actually the reason for the success of the carbon detection experiments because, in principal, you can sample only heteronuclear chemical shifts in all the dimensions of the experiments, maximizing the dispersion of the cross peaks.
Another peculiar property is the very fast chemical exchange processes of amide protons with the solvent. It is better if you try to approach physiological conditions such as neutral pH and physical temperature.
Often, if amide protons are exposed to the solvent, they do broaden out beyond detection because of these fast exchange processes with the solvent itself. Again, in these conditions, carbon detected experiments become a very valuable tool to recover information that, otherwise, would usually just be lost when performed with amide proton detected experiments.
This effect of exchange, on the other hand, tweaking a bit the conditions, can also be used to speed up longitudinal recovery of amide protons. Therefore, we can exploit all the tricks that have been recently proposed in the literature, to reduce the duration of NMR experiments; we can reduce the time we need to wait between performing two successive experiments. This is the so-called fast method and that can also be applied to the study of IDPs.
Finally, probably the measure or important ingredient to be able to address the study of more and more complex IDPs, is that of using multi-dimensional experiments with dimensionality higher than 3. This way, we can spread out more and more the cross-peaks in more and more dimensions in order to reduce the chances of accidental cross-peak overlap.
For this, a variety of nice approaches have been proposed in the literature. I think the ones that really stimulated the practical use of these experiments were those that gave the user an easy way to visualize these highly dimensional objects.
I should admit that, personally, they were a bit scary for me; I'm used to dealing with 3D. Looking at 3D spectra, I was reassured, but when I was thinking about looking at 4D or 5D, initially I was pretty skeptical.
It was also stimulated by a bio-NMR access project that we focused on in recent years and also thanks to, for example, collaborations with Bernhard Brutscher and Wiktor Koźmiński that we have now set up a complete suite of NMR experiments that are either based on carbon detection or on proton detection. That now enables us to focus on proteins as complex as 400 amino acids, for example.
How can IDPs affect the prevalence of genetic disorders? What role do they play within the human proteome and diseasome?
We started working on IDPs by looking at alpha-synuclein, which is very interesting. It's involved in the onset of neurodegenerative diseases but I must admit, in our hands, it was just the PET IDP protein to be used as a standard sample for the set-up of NMR experiments and to try out and develop new experiments.
We want to use all these experiments to characterize new proteins. One of the general fields where they can be applied is the one we started from, which is viral proteins. Viral proteins have a fairly small genome, so they need to make good use of their information in terms of encoding specific functions associated with different bits of amino acids.
The idea of disorder and of small interaction motifs encoded by just a few amino acids, now generally referred to as SLiMs (Short Linear Motifs), is a very appealing strategy for a virus to use a fairly compact amino acid sequence and encode this sequence with a variety of different functions.
Generally, intrinsic disorder is very abundant in viral proteins and we have been studying two proteins, one called E7 from the human papilloma virus – an oncogenic virus - and one called E1A from adenovirus, a homologous protein to E7. These are two proteins expressed in the early phases of viral infection.
It's amazing that just these quite compact polypeptide chains (one is less than 100 amino acids, the other less than 300) are able to engage in a huge variety of interactions with proteins of the host cell.
To quote the expression from one the key papers on this subject, they are able "to hijack cell regulations." This is one of the topics that we've been focusing on and now we are about to publish the first results in this field.
Another area where these methods can become really useful is the study of the so-called flexible linkers. We focused on this subject in collaboration with Peter Tompa. Often, when we look at complex molecular machineries, they are often constituted by several modules that are folded, connected by these flexible linkers.
This is the case with many transcription factors, for example. One of them, CBP, is about 2,500 amino acids and the folded domains have been characterized over the years, which has explained a lot about the function of the protein.
On the other hand, about half of the polypeptide chain is not folded, so somehow it is in an intrinsically disordered state. It would be quite a waste if nature had used half of the amino acid sequence to behave just as ropes between folded units.
Therefore, one other project we are focusing on is the characterization of the intrinsically disordered fragments of this protein, to try and figure out if they really are just ropes or if there are other functional elements encoded in these flexible bits and pieces of proteins.
However, this appears to be a very general property of complex proteins and this leads me to the final topic where I think NMR can make a nice contribution. That comes from the study of small intrinsically disordered fragments of another important transcription factor, the androgen receptor, a topic we focused on in collaboration with Xavier Salvatella.
This is also a complex protein, for which we have focused on the first 150 amino acids. It seems not so interesting, just a small bit of a complex protein, but, this is the part where there is a chance that the five glutamines expand and increase in number, when the disease progresses.
These diseases are called polyQ diseases because these fragments in this first bit that we are studying, when the disease progresses, become characterized by 20, 25, 30 or 35 glutamines in a row. These bits and pieces do not crystallize, so there is not much known about their high resolution properties and how these are linked to the onset of disease.
If you think in terms of using NMR to look at a part that has 30 Qs in a row, at the beginning, we thought we would never be able to characterize it. It’s a highly repetitive sequence.
In this context, the methods that have recently developed that focus on IDPs to achieve high resolution and characterize flexible systems, allowed us to characterize this first segment, which included these 25 glutamines in a row. You can then use these data to try to explain the reasons these amino acids then cause several diseases.
A well-known example of one of these polyQ diseases is Huntington’s disease. The molecular origin is the same; a fragment of a few Qs that then becomes longer and longer. The protein we focused on is the N-terminal fragment of the androgen receptor.
The malfunction causes the disease called SBMA (spinal and bulbar muscular atrophy), which is a rare disease. It's known to be related to the oligomerization of these polyQ fragments. The more Qs there are, the more these proteins tend to aggregate instead of remaining in their physiological native state and that gives rise to the disease.
This is, again, a field where NMR can provide really tremendous insights because, of course, it's disordered proteins and thanks to the recently developed methods, one can overcome the limitations deriving from high overlap of these highly repetitive sequences.
We have been focusing on this project of carbon detection since 2003, when we got the first probe with improved sensitivity for carbon detection. We started by looking into this topic and, in small steps, arrived at developing a whole set of carbon detected multi-dimensional experiments that actually are in the Bruker release.
This is in collaboration with Wolfgang Bermel, Rainer Kümmerle, who have always been part of this project in a team with my colleague Roberta Pieratelli. I have shared all the work I've been talking about with her, so I should give her credit as well.
While moving our careful attention to IDPs, we also focused on methods that were not only based on carbon detection, but tried to combine the most useful methods based on amide proton detection and also eventually, on Hα proton detection that is not affected by exchange.
Now, I think a complete suite of multi-dimensional experiments is available and can be easily implemented with recent software to really provide a lot of insight into the study of IDPs. It allows us to study fairly complex systems.
What does the future hold for your research and research into IDPs? How will you use Bruker NMR equipment in your future research?
When we started on this topic, the largest systems characterized were 100 to 150 amino acids long, whereas, now, there are several examples of IDPs as long as 400 amino acids that show that this can really be characterized at atomic resolution. And so, why not?
Maybe, when we have the 1.2 gigahertz, we will manage to think about characterizing IDPs as complex as, perhaps, a 1000 amino acids and contribute more and more to this exciting field.
Since we have been devoting so much attention to the topic of carbon detection, it would be really great to see some planning of some developments at 1.2 gigahertz because the increased magnetic field would be an obvious benefit.
It would be very interesting to see how the carbon detected experiments perform at higher fields. Carbon detection has a lower sensitivity with respect to proton detection, but also has some advantages. For example, you don't need to suppress the solvent signal.
There are not big detrimental effects from the higher ionic strength generally used to study some IDPs and so maybe, it would be an easy way for Bruker to try the new technologies and see if we can make, in the next 10 years, another jump of a factor of 10, which would, of course, be very appreciated.
Where can readers find more information?
An overview on my research and teaching activity, my CERM page
Bruker’s NMR technology and applications
About Dr Isabella Felli
I'm Isabella Felli. I've been working here at CERM since my undergraduate thesis in 1993 under the guide of Professor Ivano Bertini, my mentor when I started in the field of NMR. He actually created at CERM a very stimulating scientific environment and started this very large research infrastructure that now has eleven instruments and has been providing access to external users all over the world ever since, mainly to Europe, but all over the world.
I'm very proud to have a chance to use all the beautiful instruments that are available here in Florence and, also, to be involved in a variety of different projects focusing on the development of NMR methods. I am also stimulated by all the feedback we get from the external users.