Please can you give a brief overview of your work and the techniques you use?
I am a professor in the Department of Chemistry and Biochemistry at the University of Delaware. I am a physical chemist and I work in the field of biophysical chemistry and solid-state NMR spectroscopy.
My research lab studies several classes of systems. We are mostly interested in looking at large protein assemblies to understand their structure, dynamics and how their properties relate to their malfunction in disease.
Tatyana Polenova - Studying Protein Dynamics in Disease with NMR
Tatyana Polenova - Studying Protein Dynamics in Disease with NMR from AZoNetwork on Vimeo.
We work on two classes of systems; one is proteins that come from the cytoskeleton, which are associated with microtubules and actin, and the other is proteins that come from the HIV-1 virus. Both of those classes of proteins are related to disease.
In another effort, we are interested in looking at the role of transition metals that contain half-integer quadrupolar nuclei.
We look at half-integer quadrupolar metal sites in proteins and inorganic materials. This research requires a lot of different complementary techniques, and at the center of it is NMR spectroscopy.
We mostly use solid-state NMR magic angle spinning methods, but we also carry out a lot of computational studies. We perform quantum chemical calculations of electronic structure and, in collaboration with other groups, we do molecular dynamics simulations.
What are the key objectives of your research with regards to the HIV virus and cytoskeletal proteins?
Our research is based on two pillars. One is looking into the systems, into the biology; the other is using the latest available technology and developing technology that enables us to do that.
At the moment, the current methods are not always sufficient to answer the biological questions of interest. For examples, we know that HIV-1 is a global pandemic and despite years of research by many investigators, there is still no cure. We ask ourselves how we can go about it in our little corner of structural biology.

We are part of the Center for HIV Protein Interactions which is based at the University of Pittsburgh and led by Angela Gronenborn.
This center brings together people from different disciplines including virology, biochemistry, as well as structural biology in order to answer challenging questions about the HIV-1 virus.
At the center, experimental methods, such as NMR spectroscopy, cryo-EM, and X-ray crystallography, are integrated with computational techniques, to gain in-depth understanding into the virus structure and function.
We want to know how the structures of the virus and the interactions with proteins that comprise the host cell, play a role in the disease. By understanding the basic science, we hope that we will be able to contribute to finding a cure for the virus.
What is most important is that this kind of work is not limited to a particular class of system. For example, methods that we use and develop to look at the HIV-1 assemblies are applicable to understanding other types of viruses and bacterial pathogens. The methods that we develop and use are very broad in this sense.
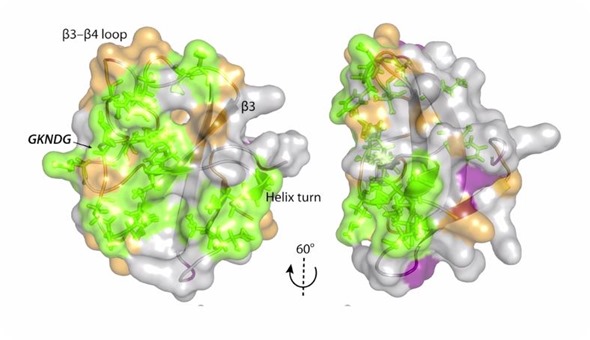
To give you another example, I mentioned proteins that are part of the cytoskeleton, which bind to microtubules and actin. It turns out that if there are mutations in these motor proteins, such as those in kinesin and dynein/dynactin complex, disease can occur.
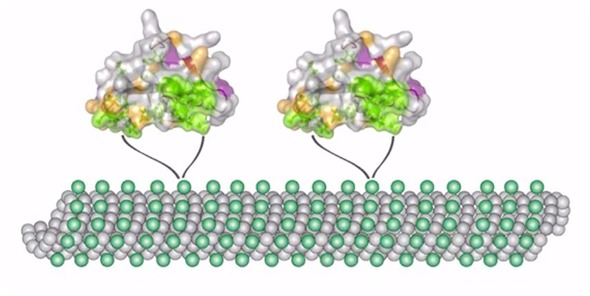
We need to understand what roles these mutations play in changing the structure. However, it turns out knowing the structure alone does not provide a full picture. We also need to know how molecules move when they are bound to these kinds of systems.
It turns out that structure and motion function in concert to either produce a healthy response or, unfortunately, cause disease.
Our research tries to find the link between this atomic-level structure and dynamics and how diseases occur, whether they are based on pathogen invasion, as is the case in HIV, or whether they are based on genetic modifications of our own proteins.
Are you expecting any major developments in technology that would really help to drive your research in the future?
Technology is the "bread and butter" of our research. We are dependent on physical methods to be able to look at challenging biological systems.
Without the latest technological developments, we would be nowhere. This is not only the case in my own lab; it is true of anyone who is interested in understanding large biological systems.
NMR spectroscopy is the main method that we use. I will talk a bit about what I think would be important additions to the arsenal of tools that we already have. I also have some thoughts on other techniques which might be important.
For the field of NMR spectroscopy, I would say that the most important development could and should be assessing higher and higher magnetic fields.
Companies such as Bruker and others, as well as national laboratories are pioneers in this.
We hope as a community of researchers, that this quest will continue because our own experience has been that when we work on large biological systems, we are currently limited by the magnetic fields that we can attain, both in terms of sensitivity and resolution. I would say this is the number one priority.
The number two priority is this real need for technological development, which in my opinion is a game changer, at least for some ranges of systems. This has to do with the accessibility of ultra-fast magic angle spinning methods.
This allows us to get ever-increased resolution and also sensitivity, so we can work with very small amounts of samples. This is important because many biological systems cannot be made in large quantities.
Also, given the complexity of the programs, we want to have narrower and narrower lines and it turns out that ultra-fast magic angle spinning in conjunction with high magnetic fields, would open doors for us to be able to do that.
A third development that has emerged where the level of interest is now surging, is dynamic nuclear polarization (DNP). This is yet another area which is really exciting for us because we can attain very large sensitivity enhancements.
It was pioneered by a number of people in the field, including Bob Griffin and implemented by Bruker in the form of commercial instruments, which is very important for the community because we can now have easy access to this kind of instrumentation.
The reason why DNP might provide breakthroughs is because we would be able to work on natural abundance complex systems.
Another factor that is an extreme challenge at the moment is the throughput. NMR is an inherently low throughput method. Unfortunately, we cannot compete with X-ray crystallography in terms of how fast we can deliver information.
On the other hand, we provide an exquisite level of detail, not only on the static structures, but also on the motional characteristics of particular systems of interest and molecular mechanisms and chemistry, which is quite unique to magnetic resonance.
How can we streamline the throughput problem? In my opinion, one area that needs development, is being able to streamline the flow from the data collection to the structure.
That requires a lot of work in computation. A stumbling block in the field of NMR spectroscopy is how to assign resonances efficiently. There needs to be a lot of software development.
How important is the integration of different methods?
Another very important aspect is the fact that in order to get the most detailed picture, we need to integrate many methods.
That has become increasingly clear over the past several years. NMR cannot function alone and nor can X-ray crystallography, theoretical chemistry or theoretical biophysics.
To be able to study complex biological systems, we need to integrate experiment and theory. There are really exciting developments in terms of these integrated techniques where NMR can provide information at atomic resolution on the details of structure and dynamics.
In parallel, cryo-electron microscopy provides lower-resolution picture about the envelope of large particles. NMR and cryo-EM data are then integrated through computation to give an exquisitely detailed picture about the biological assemblies of interest.
I would say that every scientist’s dream is probably to be able to peer into living cells with atomic resolution. How we achieve that is probably a question for all of us in the field, as well as for the young generation of scientists who are in the field already or are entering the field right now.
What do you hope the future holds for NMR?
In the field of NMR, what is extremely important is the growing development of ultra-high field magnets, which are one of the main pre-requisites for our ability to conduct research on complex biological systems.
The scientific community needs to have access to these kinds of infrastructures. The only way to do that effectively is arguably through a shared network of facilities.
No single lab could afford the expensive instruments which cost many millions of dollars, but when we build networks, that allows us all to get access to the state-of-the-art instrumentation, which is maintained in peak performance.
We can also collaborate within these networks, not only on technologies, but in terms of sharing ideas and attracting people from other disciplines who are not magnetic resonance researchers.
They can come and learn what NMR is about and bring their systems, to elevate the level of the problems and the questions that we are currently asking.