During high volumes of drug screening, different types of assays have been shown to be useful for detection. But, toward the end of these drug screenings, the positive hits have to be confirmed in living cells related to disease, for example, iPSC derived cell lines, cardiomyocytes, or primary cultures of neurons.
In this context, most of the assays employed in HTS are not sufficient because they cannot be targeted to a specific cell population. Additionally, such assays are usually end point lytic assays and/or need a substrate that has to be exogenously applied.
Montana Molecular has designed powerful, genetically encoded biosensors to meet the requirements for methods that can detect signal transduction cascades in living cells. As a proof of concept, this article shows how the CLARIOstar® microplate reader was used for detecting Gi and Gs signaling in HEK293 cells.
Assay principle
Biosensors from Montana Molecular are housed in a modified baculovirus, which has been optimized for expression in mammalian cells (BacMam). A schematic diagram of the mechanism of cAMP assay is shown in Figure 1.
In order to trigger changes in the fluorescence of a fluorescent protein, the cAMP assay uses cAMP dependent conformational changes in the EPAC cAMP binding domain. Genetically-encoded biosensors can be spectrally resolved in the living cells, and are available in either green or red version.
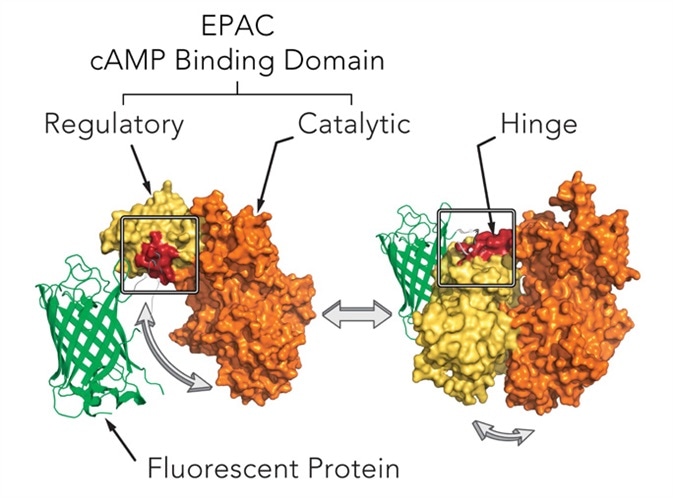
Figure 1. Mechanism of cAMP assay. Image credits: BMG Labtech.
The biosensors can decrease or increase in fluorescence. When cAMP production increases, upward cADDis increases and when cAMP production increases, downward cADDis decreases, allowing background fluorescence to be easily separated from the fluorescent assay. In this analysis, results were demonstrated from both the Downward cADDis and the Upward cADDis biosensors (Figure 3).
Gs and Gi detection using cADDis1 cAMP sensor
In Figure 2, the schematic diagram shows how the D2 receptor-coupled Gi decreases in adenylyl cyclase (AC) activity and the ß2-AR-coupled Gs increases in AC activity. Assays that are capable of detecting these signaling events on the CLARIOstar® microplate reader were validated.
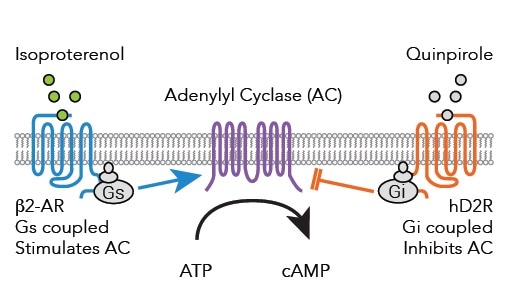
Figure 2. Signaling pathways from Gs and Gi coupled receptors to adenylyl cyclase after binding of either Isoproterenol or Quinpirole. Image credits: BMG Labtech.
Materials and methods
The following materials and methods were used:
- HEK293 cells
- Greiner 96-well F bottom plates
- Montana Molecular cADDis sensors
- BMG LABTECH’s CLARIOstar® microplate reader
Sensor transduction
HEK293 cells were plated in 96-well plates after being transduced in suspension with BacMam carrying the specified sensors. Sensor was expressed by the cells for a period of 24-28 hours, and the media was replaced with PBS. The cells were left undisturbed at room temperature for 30 minutes before the experiment.
Instrument settings
|
Detection Mode
|
Fluorescence Intensity (Bottom)
|
|
Detection Method
|
Plate Mode, Kinetic
|
|
Scan mode
|
Orbital Averaging
|
|
Scan diameter mm)
|
4
|
|
Gain / Focal height
|
Adjusted prior to test
|
Optical settings
|
Excitation
|
F 482-16
|
|
Dichroic
|
LP 504
|
|
Emission
|
F 530-40
|
Results and discussion
Isoproterenol stimulated Gs detection using cADDis cAMP sensors
The CLARIOstar® microplate reader was used to assess the cADDis detection. This was done by using the well-defined response of HEK293 cells to Isoproterenol as the background. To assess baseline fluorescence, cells were observed for 2 minutes (Figure 3) and then treated with 30 μM PBS or Isoproterenol before again observed for another 10 minutes.
Using a downward or upward green-cADDis sensor, the assay performance was found to be robust based on the Z’ factor of more than 0.85. The downward cADDis sensor somewhat shows a better performance (Z’ = 0.941) in part, because the sensor’s signal is reverse of any background fluorescence.
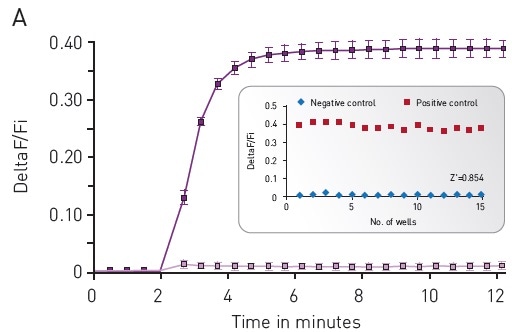
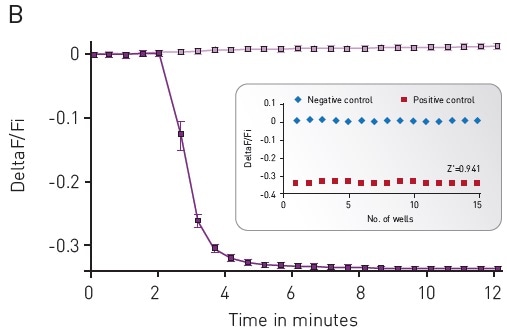
Figure 3. Green-cADDis sensor kinetics for (A) upward sensor and (B) downward sensor in response to isoproterenol Mean +/- std; n = 15 wells. Inset: Gs assay performance in 96-well plate. For (A) Upward sensor Z’ factor is 0.854. For (B) downward sensor Z’ factor is 0.941. Image credits: BMG Labtech.
Forskolin-free Gi detection using cADDis sensors
The cADDis sensors have been adapted for detecting Gi signaling in a forskolin free assay. The effect of Gs at adenyl cyclases is increased by forskolin and directly affects the ion channels.2
It is not recommended to use forskolin in the cADDis assays because it affects biological activity. Forskolin will exaggerate the response in the presence of GPCR with basal Gs activity, and the obvious EC50 of agonists at the Gs-coupled receptor is shifted.
This makes agonists appear more potent than they actually are. With regard to Gi-coupled receptors, the effect of forskolin on Gs stimulation makes it relatively hard to inhibit adenyl cyclase.3
Using co-expression of a constitutively active Gs mutant, cAMP basal levels are increased and so enables real time, kinetic measurements of Gi signaling in the living cell.
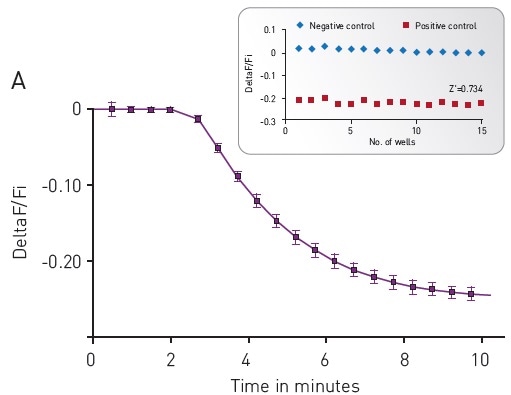
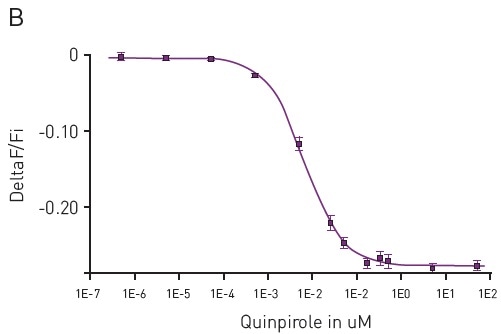
Figure 4. (A) Kinetic measurements of D2 mediated Gi signaling using green—cADDis sensor and the D2 specific agonist Quinpirole. Mean +/- SEM; n = 12 wells. Insert, Gi assay performance in 96-well plate. Z’ factor is 0.741. (B) Dose response to Quinpirole. EC50 is 5.7 nM. Mean +/- SEM. n = 6 wells / condition. Image credits: BMG Labtech.
Conclusion
The genetically encoded assays from Montana Molecular can be effectively used with the CLARIOstar® microplate reader. Dose and kinetic response data for Gs and Gi signaling in living cells were achieved. The combination of sensors and reader resulted in robust Z’ values.
Acknowledgements
Produced from materials originally authored by Paul Tewson1, Scott Martinka1, Shane Tillo1, Thom Hughes1, Anne Marie Quinn1, Carl Peters2 from:
1 Montana Molecular, Bozeman, MT
2 BMG LABTECH Inc., NC, USA
References
- Tewson, P.H. et al. (2015) J. Biomol. Screen. 21(3):298-305.
- Hoshi, T. et al. (1988) Science 40(4859): 1652-1655.
- Dessauer, C.W. et al. (1998) J. Biol. Chem. 273(40): 25831-25839.
About BMG Labtech

BMG LABTECH has been committed to producing microplate readers for more than twenty years. By focusing on the needs of the scientific community, the company’s innovative microplate readers have earned the company the reputation of being a technology leader in the field.
BMG LABTECH has developed a wide range of dedicated and multi-mode microplate readers for life sciences applications and high-throughput screening.
All BMG LABTECH microplate readers are "Made in Germany" and are conceived, developed, assembled, and tested entirely at our headquarters in Germany.
Since our establishment in Offenburg, Germany in 1989, BMG LABTECH has expanded to offer a worldwide sales and support network with offices in the USA, UK, Australia, Japan and France. Our subsidiaries, regional offices and distributors are committed to bringing you innovative microplate reader technology with the quality and reliability you expect from a German company.
Our staff includes engineers and scientists from the fields of biology, biochemistry, analytical chemistry, and physics.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.