Understanding the kinetics of protein folding that are linked to protein stability is essential to gain knowledge about the structure, energetic and dynamic properties of proteins. Carrying out Ф-value analyses is advantageous, but data collection is laborious for such analyses. Moreover, in most cases, Ф-value analyses are not suitable to high throughput.
This article discusses the capability of the Omega microplate reader to measure tryptophan (Trp) fluorescence and adapt it for kinetic studies By leveraging the reagent injection option, the method that directly measures the unfolding rates of proteins.
Assay principle
Two assay methods were developed. Figure 1 shows the kinetic protein unfolding assay, which involves the injection of the denaturant and subsequent monitoring of the fluorescence emitted by the intrinsic tryptophan residues that reflects protein unfolding. Moreover, Omega’s injection at the point of measurement feature enables the user to record maximum amounts of signal information.
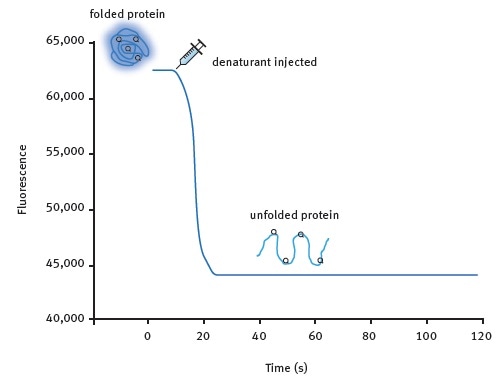
Figure 1. Kinetic protein unfolding assay principle. The folded form of the protein exhibits relatively high intrinsic Trp fluorescence. Injection of a denaturing solution leads to protein unfolding and a decrease in fluorescence that can be monitored in real-time for many proteins. Image credits: BMG Labtech.
A limited proteolysis assay approach was developed for proteins possessing an unfolding kinetic constant of more than 0.01s-1. Trp (tryptophan) fluorescence was monitored over time by manually adding varying concentrations of thermolysin or trypsin to the protein.
Materials and methods
- 96-well Quartz microplates from Hellma
- POLARstar Omega microplate reader from BMG LABTECH
Full experimental details can be read in detail in Wang et al.1 . Proteins analyzed were AbpSH3 (the src homology 3 domain from Actin Binding Protein 1), and its mutants AbpSH3 E7L, AbpSH3 V21K and AbpSH3 Triple, which carries the mutations E7L, V21K, and N23G. An AbpSH3 – ArkA (Actin Regulating Kinase SH3 binding peptide A) hybrid called hybrid long linker (HLL) was also tested.
Omega instrument settings
Kinetic protein unfolding assay
|
Measurement type
|
FI (Bottom)
|
|
Measurement mode
|
Well Mode
|
|
No. of cycles
|
1000
|
|
Cycle time
|
120 ms
|
|
Filters
|
Ex: 280-10 / Em 330-10
|
|
Gain
|
1400
|
|
Injection volume
|
4.0 – 92.0 μl (4.0 μl increments)
|
Limited proteolysis unfolding assay
|
Measurement type
|
FI (Bottom)
|
|
Measurement mode
|
Plate Mode
|
|
No. of cycles
|
1000
|
|
Cycle time
|
73 s
|
|
Filters
|
Ex: 280-10 / Em 330-10
|
|
Gain
|
1400
|
Results and discussion
Figure 2 shows the representative data obtained for the wild-type AbpSH3 protein with the Omega plate reader. The data shows the unfolding curves created at various concentrations of the denaturant guanidine.
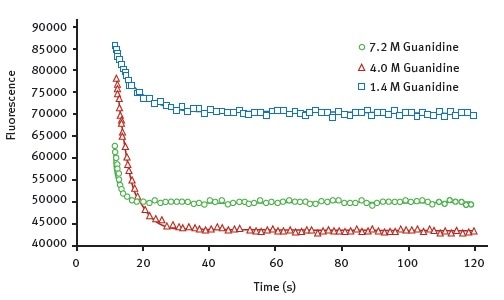
Figure 2. Kinetic traces for the protein unfolding of AbpSH3 WT. The final concentration of guanidine, injected at 12 seconds is indicated. The data is fit to an exponential decay equation. Image credits: BMG Labtech.
Using these data, it is possible to determine Chevron plots of the unfolding in order to determine unfolding constants listed in Table 1. It is to be noted that the values measured for AbpSH3 and its mutant forms agree well with the values previously published in the literature2,3. This means that this assay method is as good as stopped-flow approaches.
Table 1. Summary of kinetic unfolding data
|
|
ku exp. (s-1) X 103
|
ku literature (s-1) X 103
|
mu exp. (kJ mol-1 M-1)
|
mu literature (kJ mol-1 M-1)
|
|
AbpSH3 WT
|
71.0 ± 5.4
|
65.8 ± 5.3
|
0.92 ± 0.06
|
0.83 ± 0.04
|
|
AbpSH3 E7L
|
7.2 ± 0.5
|
4.8 ± 1.6
|
1.09 ± 0.01
|
0.96 ± 0.10
|
|
AbpSH3 V21K
|
48.5 ± 10.5
|
44.8 ± 4.3*
|
0.69 ± 0.05
|
0.56 ± 0.03*
|
|
AbpSH3 Tri.
|
2.9 ± 0.6
|
4.7 ± 3.6
|
1.01 ± 0.01
|
0.70 ± 0.20
|
|
HLL
|
4.2 ± 0.8
|
n/a
|
1.07 ± 0.01
|
n/a
|
* Experiments used urea instead of guanidine as denaturant
The results obtained for the limited proteolysis unfolding assay using HLL are shown in Figure 3, they illustrate monitoring of protein unfolding in the presence of trypsin.
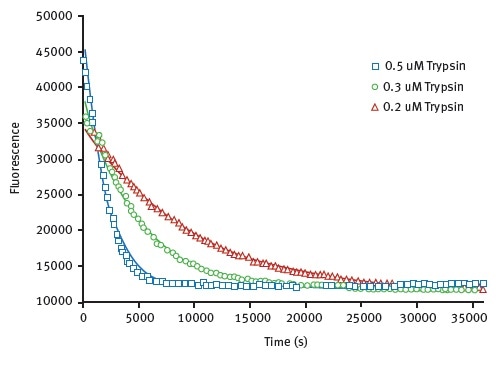
Figure 3. Unfolding kinetic constant derived from limited proteolysis of HLL. Representative raw data from proteolysis of HLL at 3 different trypsin concentrations. Degradation is followed at 330 nm. The data is fit to exponential decay curve. Image credits: BMG Labtech.
Using a Michaelis-Menten type equation with this data, unfolding constants for trypsin was found to be 0.0023s-1. A value of 0.0042s-1 can be seen in Table 1.
These results confirm that this complementary approach provides a viable alternative method for fast unfolding proteins. Therefore, the Omega enables both the chemical denaturation method and limited proteolysis methods to achieve reliable, high throughput kinetic protein unfolding measurements.
Conclusion
The kinetic protein unfolding assay is advantageous in many ways over previously explained approaches. A shorter dead-time is of one of the advantages of this assay, enabling the monitoring of proteins unfolding relatively quickly.
The capability of the Omega to collect readings every 120 ms is another advantage of this assay. Both assays discussed in this article can be carried out in a relatively low volume of around 100 µl protein concentration that will save samples.
Acknowledgments
Produced from materials originally authored by Qiang Wang, Zoey Sharp, Nicklas Waterhouse, Matt Dominguez and Elliott J. Stollar Dept. of Physical Sciences, Eastern New Mexico University, USA.
References
- Wang Q. et al. (2016) Development and Application of a High Throughput Protein Unfolding Kinetic Assay PLoS ONE 11 (1):e0146232.
- Rath A and Davidson AR (2000) The design of a hyperstable mutant of the Abp1p SH3 domain by sequence alignment analysis Protein Sci. 9(12):2457- 2469.
- Maxwell KL et al. (2005) Protein folding: defining a “standard” set of experimental conditions and a preliminary kinetic data set of two state proteins Protein Sci. 14(3):602-616.
About BMG Labtech

BMG LABTECH has been committed to producing microplate readers for more than twenty years. By focusing on the needs of the scientific community, the company’s innovative microplate readers have earned the company the reputation of being a technology leader in the field.
BMG LABTECH has developed a wide range of dedicated and multi-mode microplate readers for life sciences applications and high-throughput screening.
All BMG LABTECH microplate readers are "Made in Germany" and are conceived, developed, assembled, and tested entirely at our headquarters in Germany.
Since our establishment in Offenburg, Germany in 1989, BMG LABTECH has expanded to offer a worldwide sales and support network with offices in the USA, UK, Australia, Japan and France. Our subsidiaries, regional offices and distributors are committed to bringing you innovative microplate reader technology with the quality and reliability you expect from a German company.
Our staff includes engineers and scientists from the fields of biology, biochemistry, analytical chemistry, and physics.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.