There are many types of experimental methods that often use normalization to fix the differences induced by factors other than what is immediately being analyzed. In particular, normalization can be very useful in luminescent genetic reporter experiments, because transient transfection techniques that are frequently used can cause variability from several sources, like those provided in Table 1.
When a normalization step is included, variability is reduced, data comparisons are made easier, and statistical importance and confidence in the data are improved. While several techniques are available to account for the normalization process, this article mainly focuses on data analysis and experimental optimization when utilizing co-transfected control reporters.
Table 1. Potential sources of variability in transient transfection methods
|
Source of Variability
|
Potential Causes
|
|
Starting cell number
|
Pipetting variation, problems with clumping/dispersion
|
|
Transfection efficiency
|
Difficult to transfect cells, suboptimal transfection, variable cell density.
|
|
Ending cell number
|
Cytotoxic effect of treatment, detached cells lost in media transfers or washing steps.
|
|
Position in multiwell plate
|
Edge effects brought about by differences in heat distribution and humidity across a plate.
|
Available options for normalization
Normalization to cell health (viability), normalization to total protein content, or normalization to a co-transfected internal control reporter constitute the standard methods used for normalization of genetic reporter data.
For instance, normalization to total protein content using a Bradford assay not only controls the variation in total cell number but may also be utilized when working with steadily transfected cells where variability sources are much more restricted. Protein assays, however, are the least facile option (for example, these assays cannot be multiplexed with reporter assays) and hence, are not discussed in this article.
Multiplexing with markers of cell health
Multiplexing a reporter assay with a compatible cell viability — although not essentially used for normalization — provides a way to gain a better insight into reporter gene expression with regard to cell health.
A nonlytic, fluorescent viability assay, for example, can be carried out upstream of a luminescent reporter assay which will allow sequential analysis of reporter expression as well as viable cells from the same well.
In a single-reporter assay or dual-reporter assay using two experimental reporters, a cell health assay will make it easy to understand the data and at the same time, will account for reduced reporter activity induced by the toxicity of compound treatment, as shown in Figure 1.
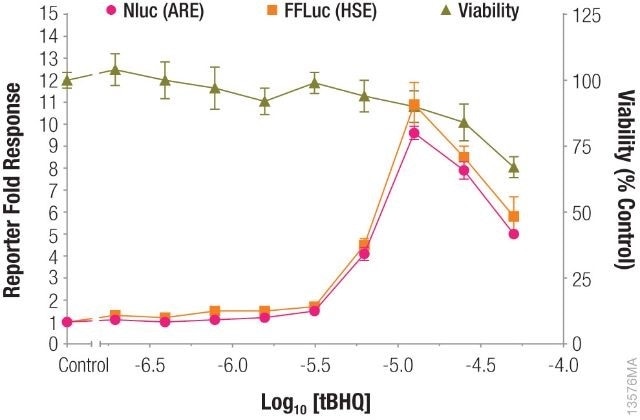
Figure 1. Measuring antioxidant response element (ARE) and heat shock response element (HSE) responses followed by cell viability from the same sample. Image credit: Promega
pGL4.41[luc2P/HSE] and pNL[NlucP/ARE/Hygro] were used to transfect HepG2 cells, which were subsequently treated with tBHQ. After incubating the HepG2 cells overnight, cell viability was measured using the CellTiter-Fluor™ Cell Viability Assay, and ARE and HSE responses were measured using the NanoDLR™ assay.
Working with internal control reporters
The best way to regulate the variables introduced in transfection-based experiments is normalization to an internal control reporter. In this approach, a constitutively expressed control reporter vector is used which is then co-transfected with the empirical vector.
A dual-reporter assay (Table 2) is used to sequentially measure the luminescence of both vectors, and the “Experimental Reporter Activity/Control Reporter Activity” is calculated to obtain a normalized ratio for each well.
This technique accounts for variability from one well to another caused by variations in transfection efficiency, number of viable cells, cell number, or edge effects induced by the plate position.
Shown in Figure 2 is the difference in coefficients of variation (CV) for data obtained from one single-reporter assay without normalization against the data acquired from a dual-reporter assay such as Dual-Glo® or NanoDLR™, with normalization. Data normalization for each well not only fixes the variability but also cuts down the overall CVs acquired to a large extent, thereby enhancing data quality.
Table 2. Comparison of Dual-Luciferase Reporter Assays
|
|
Dual-Luciferase® Assay
|
Dual-Glo® Assay
|
Nano-Glo® Dual-Luciferase® Assay
|
|
Format
|
Non-Homogeneous
|
Homogeneous
|
Homogeneous (Also compatible with prelysis)
|
|
Sample Process
|
Bench-Scale
|
Bench to Batch
|
Bench to Batch
|
|
Number of Steps
|
5
|
2
|
2
|
|
Sensitivity
|
Higher
|
Lower
|
Higher
|
|
Firefly Signal Half-Life
|
~ 9 minutes
|
~ 2 hours
|
~ 2 hours
|
|
Renilla Signal Half-Life
|
~ 2 minutes
|
~ 2 hours
|
NA
|
|
NanoLuc® Signal Half-Life
|
NA
|
NA
|
2 hours
|
|
Precision
|
High
|
High
|
High
|
|
Cell Lysis Time
|
~ 15 minutes
|
~ 10 minutes
|
~ 3 minutes
|
|
Recommended Experimental reporter
|
Firefly
|
Firefly
|
Firefly or NanoLuc®
|
|
Non-Homogeneous: Lysate created before reagent addition.
Homogeneous: Reagent added directly to cells in culture.
NA: Not applicable.
|
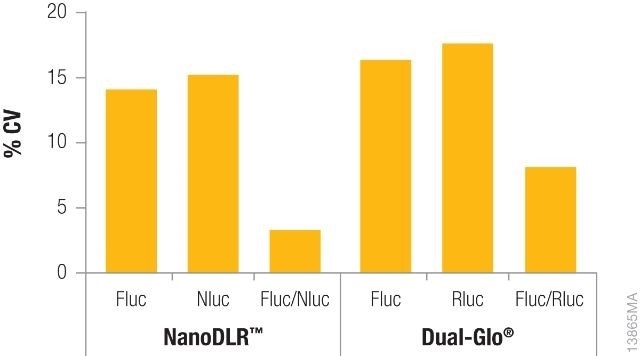
Figure 2. Comparison of coefficients of variation (CVs) obtained from a single reporter read vs. normalized data using either the NanoDLR™ or Dual‐Glo® assays. Image credit: Promega
Constitutively expressed firefly luciferase and constitutively expressed Renilla luciferase (Dual-Glo® Assay) or NanoLuc® (NanoDLR™) were used to transiently transfect the inner 60 wells of a 96-well plate of HEK293 cells for each reagent.
After overnight incubation, the media was removed, the plate was washed with PBS, and the new media was added. Firefly luciferase levels were first determined and then Renilla or NanoLuc® levels were measured using Dual-Glo® or NanoDLR™, respectively. For individual reads, CVs were determined across each plate and again determined after normalizing the data.
Optimizing your control-reporter experiments
It is important to choose a control reporter and promoter. It is also imperative to make sure that the reporter employed as the internal control is entirely different from the one employed as the experimental reporter.
This is because the activity of both reporters will be determined in the same cell lysates or cells, as indicated in Table 2. There should be enough signal from the control reporter to enable accurate and easy determination but at the same time should be low enough, so that there is no interference with the experimental reporter expression, which is discussed below in detail, and that any issues reacted to detection assay (saturation of the detection instrument or substrate depletion) is also prevented.
Providentially, this leaves a very broad range. The overall signal obtained is further contributed by the luminescent reporter as well as the promoter driving its expression. From each reporter/promoter combination, the relative luminescence can differ based on the specific type of cells being used, as illustrated in Figure 3. As a result, it is vital to consider and optimize co-transfection conditions for specific experimental conditions.
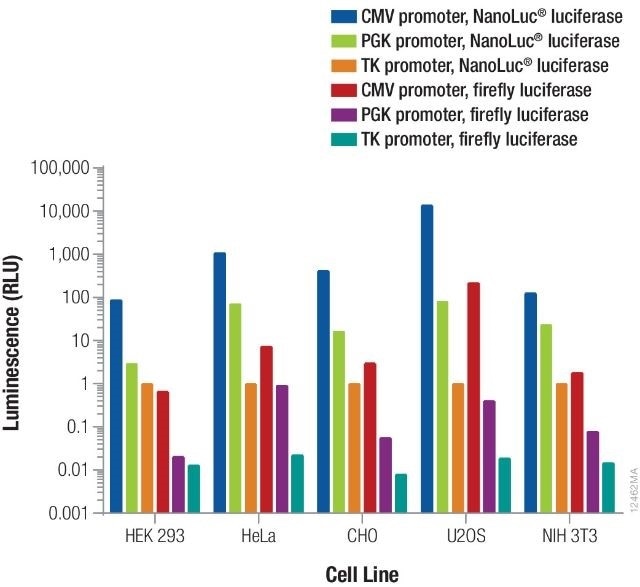
Figure 3. Relative luminescence for firefly and NanoLuc® luciferases expressed from constitutive promoters in multiple cell types. Image credit: Promega
Using different quantities of DNA containing NanoLuc® or firefly luciferase expressed from TK, PGK, or CMV constitutive promoters, cells were transiently transfected. The NanoDLR™ Assay was used to measure luminescence following 24 hours expression and the estimated relative luminescence as opposed to TK-NanoLuc® was measured for those amounts of DNA that give usable signals.
The values obtained can inform the option of control reporter construct as well as the appropriate dilution or amount of DNA that needs to be transfected to obtain the required signal.
Optimizing co-transfection conditions
In addition, the amount of the control vector contained in the co-transfection determines the expression level of the control reporter construct. It is also important to determine the optimal experimental vector:control vector ratio.
Normally, the minimum amount required will be co-transfected to provide considerable reporter activity over the background. Transcriptional squelching, and other similar obstruction with the experimental promoter, that is, trans effects between the promoters may occur due to transfection of high amounts of control plasmid.
The significance of reducing co-reporter levels is shown in Figure 4. When the Renilla co-reporter was co-transfected at a ratio of 10:1 (firefly:Renilla), the firefly luciferase signal was reduced almost by half, i.e., 50%, and at a ratio of 1:1 the same firefly luciferase signal was reduced by approximately 70%.
An interesting fact was that co-transfection of the NanoLuc® luciferase did not had had a major effect on the firefly luciferase signal and causes just 20% inhibition at 1:1 and 10:1 transfection ratios.
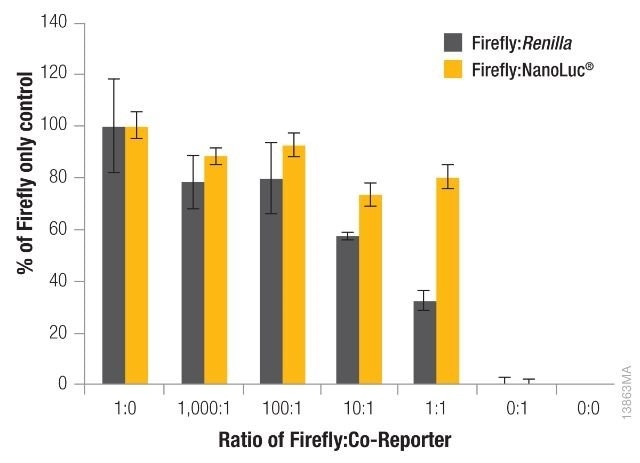
Figure 4. The effect of co-reporter expression levels on firefly signal with NanoDLR™ or DLR™ Assay. Image credit: Promega
Co-reporter (Renilla or NanoLuc®) vector mass considerably differed from 5 to 500µg, while firefly luciferase vector mass was sustained at 500ng. N = 8 for each condition. Transfection carrier DNA was added to maintain the total transfection amount at 1μg.
It is advised to first optimize the conditions for the experimental reporter to improve the amount of co-reporter required for assay conditions. Next, the amount of co-reporter required can be empirically determined by co-transfecting a differing amount of control vector to establish the least amount that gives adequate response to be confidently determined.
Shown in Figure 5 is at typical example of this kind of optimization experiment, wherein firefly levels are maintained constant as the experimental reporter, while transfecting an increasing amount of a TK-driven co-reporter, NanoLuc® or Renilla luciferase.
The above data shows that both the detection reagent and the co-reporter will determine the optimal amount of co-reporter. When Renilla luciferase is used as a co-reporter, higher co-transfection amounts would need to be used to achieve an acceptable level of signal, more so if the Dual-Glo® detection reagent is used, and extreme care should also be taken to make sure that the effect on the experimental reporter is not seen.
When Renilla luciferase is used as control reporter, adequate expression levels may be obtained by using a stronger promoter. On the contrary, a strong response is given by NanoLuc® luciferase even at the lowest co-transfection level, allowing the NanoLuc® control reporter to be applied at levels that are 10,000 times lower than that of the firefly reporter (10,000:1).
To achieve proper expression levels, it is advised to express NanoLuc® luciferase from a constitutive promoter that is not as active as a CMV promoter, such as TK or PGK. Also, it is not necessary for the signal given by the control reporter to be lower or higher than the signal given by the experimental reporter.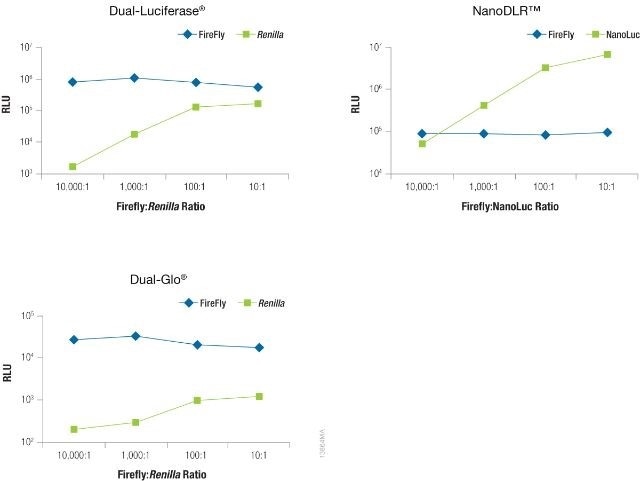
Figure 5. Raw RLUs for firefly and co-reporter (NanoLuc® or Renilla) luciferase when co-reporter DNA amount was titrated against a constant level of firefly luciferase (Fluc). Image credit: Promega
Co-reporter (Renilla or NanoLuc®) vector differed from 500pg to 50ng, while firefly luciferase vector was held constant at 500ng. N = 8 for each condition. Transfection carrier DNA was added to maintain the total transfection amount at 1μg.
Analyzing your co-reporter data
Once the normalized ratio is measured for each well, the data obtained can be directly used, or based on the experimental design, can be further examined in several ways. As an easy method of analysis, the normalized data can be averaged for each treatment and the fold change in activity between the two test groups can be determined by using the equation given below:
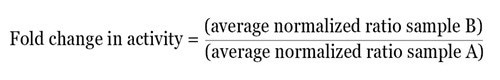
By using this method, the relative difference in activity between multiple treatments within one experiment can be obtained. From each experiment, the normalized fold change in activity can be easily averaged and statistical analysis can be carried out.
Calculating a relative response ratio
Calculating a relative response ratio (RRR) is another method. The RRR makes it possible to compare various treatments from different experiments as it gives a basis to compare treatment effects. For RRR calculation, two sets of controls on each plate have to be included — a negative control that gives basal or minimal expression and a positive control that gives the highest or maximal expression from the experimental reporter.
If both of these samples are added on each plate, then the effect of each new compound can be quantitatively assessed by its effect on the experimental reporter within the context of the negative and positive control.
For instance, when the ratio of experimental reporter luminescence against control reporter luminescence is 1.3 for the negative control, 53 for the positive control, and 22 for the experimental treatment, then it becomes possible to scale these values so that a value of 1 is assigned to the positive control and a value of 0 is assigned to the negative control. The following formula can be used to measure the RRR for each experimental treatment:
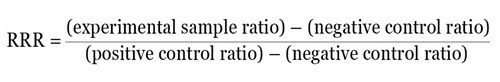
Therefore, the RRR for the above experimental treatment example would be:
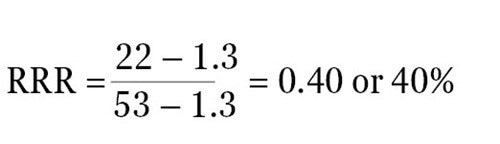
The experimental compound was found to be 40% as effective as the positive control at growing expression of the experimental reporter at this specific concentration.
It must however be noted that if the signals’ raw luminescence signal values are close to the background luminescence signal values (from media-only control wells or non-transfected cells), the luminescence values would have to be background subtracted before calculating the ratios.
Conclusion
The most effective method for reducing inconsistency in temporary transfection-based genetic reporter experiments is to include an internal control reporter. When an experiment is being designed to include a control reporter, several parameters have to be taken into account to achieve the best results.
Particular experimental conditions, i.e., the number of cells, the size of the wells, the detection reagent used, and the cells’ ability to be transfected mostly govern the determination of appropriate promoter, co-reporter, and DNA dilution for co-transfection.
While NanoLuc® luciferase is generally used a sensitive experimental reporter, it still serves as a good alternative to Renilla as an appropriate co-reporter for firefly luciferase.
The bright signal of NanoLuc® luciferase, approximately 100 times brighter than Renilla, enables it to be used at extremely low dilutions, thus reducing the effect on the firefly luciferase reporter and minimizing the total amount of DNA needed for the transfection.
Acknowledgements
Produced from materials originally authored by Chris Eggers, Brad Hook, Samantha Lewis, Carl Strayer and Amy Landreman from the Promega Corporation.
About Promega
With a portfolio of more than 3,000 products covering the fields of genomics, protein analysis and expression, cellular analysis, drug discovery and genetic identity, Promega is a global leader in providing innovative solutions and technical support to life scientists in academic, industrial and government settings.
Promega products are used by life scientists who are asking fundamental questions about biological processes as well as by scientists who are applying scientific knowledge to diagnose and treat diseases, discover new therapeutics, and use genetics and DNA testing for human identification.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and iform site visitors interested in medical research, science, medical devices and treatments.