Diffusion tensor imaging (DTI) is a newly developed magnetic resonance imaging (MRI) technique that analyzes the anatomy of nerve cells and a complex neuronal network of the brain.
What is DTI?
DTI technique was first introduced by Peter Basser in 1994. It is an improved version of conventional MRI wherein signals are solely generated from the movement of water molecules. The term ‘diffusion’ denotes random thermal motion of water molecules. In other words, DTI uses the diffusion of water as a probe to determine the anatomy of a brain network, which basically provides information on static anatomy that is not influenced by brain functions.
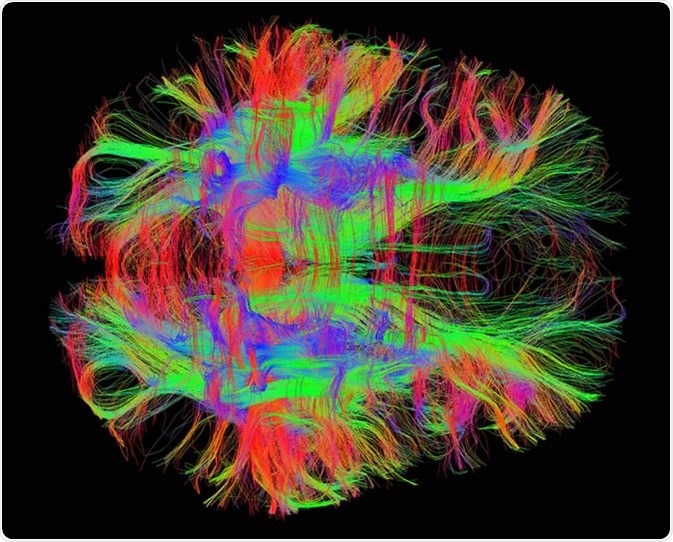
White matter fiber tracts in the adult human brain. Image Credit: Zeynep Saygin, mcgovern.mit.edu
The diffusion of water molecules in a tissue is not the same in all direction (anisotropic diffusion) due to tissue heterogeneity. This anisotropy (directional dominance of water diffusion within a region) is used in DTI to determine the nerve cell organization in the brain.
The basic principle depends on the fact that water molecules should move faster along the axon fiber instead of moving upright to the fiber because obstructions present along the fiber are comparatively lesser to restrict its movement. Based on the axonal orientation, anisotropic diffusion can produce completely new image contrast, which is very useful in visualizing important brain structures.
Diffusion Weighted Imaging
How is DTI Performed?
The analysis of water diffusion is performed by applying magnetic field gradients to generate an image that is sensitized to diffusion in a particular direction. A three-dimensional diffusion model (the tensor) is then estimated by repeating this process in multiple directions. In other words, the DTI technique involves the delivery of external magnetic pulses to impose a random phase shift for water molecules that diffuse.
This leads to a loss of signal from diffusing molecules, which subsequently creates darker volumetric pixels or voxels. For example, white matter fibers that are running parallel to the direction of the magnetic field gradient will produce a dark diffusion-weighted image for that particular direction. The diffusion tensor is then measured by comparing the signal loss with the original signal.
Two main parameters that are calculated from the tensor to define the nerve cell orientation are fractional anisotropy (FA) and mean diffusivity (MD). FA defines the degree of diffusion directionality, and MD provides the information on the average diffusivity of water.
Representation of DT Images
The information obtained from the tensor is either condensed into one number (scalar) or 4 numbers. Condensation into 4 numbers is performed to display the image with an R (red), G (green), B (blue) color and a brightness value.
Applications
Neurophysiology
DTI is a very powerful technique for investigating important aspects of basic neurophysiology as well as pathophysiological consequences associated with disorders of the central nervous system. One major advantage is that information on the orientation of nerve fiber tracts can be easily obtained through this technique.
Brain Anatomy
In addition, the spatial distribution of complex brain network can also be investigated by measuring the microscopic length scale of water diffusion. As DTI provides additional information regarding the brain anatomy over conventional MRI, it is regarded as a more sensitive tool to investigate normal brain development as well as congenital brain development disorders.
Neuropathology
Since diffusion anisotropy is significantly related to the axon myelination status, measurement of this parameter using DTI can also provide substantial insight into neuropathological conditions related to demyelination, such multiple sclerosis.
Limitations
DTI provides a thorough anatomical overview of the white matter in terms of diffusion anisotropy and fiber orientation. However, some limitations do exist related to the interpretation of the data obtained. For instance, the three-dimensional diffusion model of DTI can characterize only one fiber in a volumetric pixel; thus, a visual representation of data obtained from a region of crossing fibers can be confusing.
In other words, DTI can generate false positive or negative data in the case of brain regions that contain multiple fiber bundles oriented in different directions. Another limitation is that, since DTI is based on the diffusion of water molecules, it is not possible to differentiate axon directionalities through this technique.
Further Reading
Last Updated: Oct 11, 2018