Structured illumination microscopy (SIM) is a form of super-resolution light microscopy that enhances the resolution of the microscope. It is useful in research as it allows for high-resolution imaging of cells.
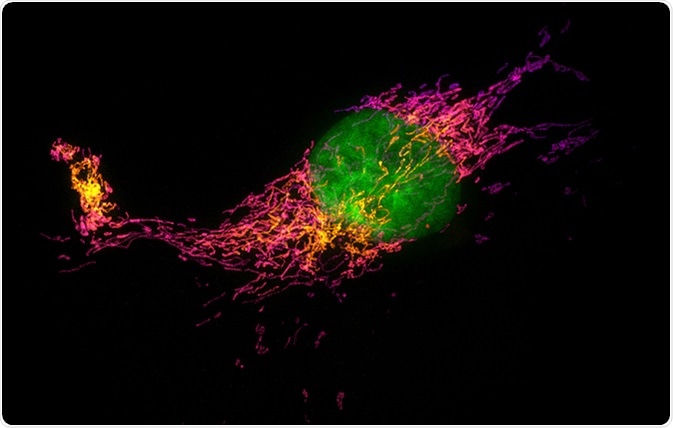
Structured illumination microscopy (SIM) images of a bovine pulmonary artery endothelial cell stained with fluorescent dyes for mitochondria and nucleus. Maximum intensity projection of a z-stack. Image Credit: Micha Weber / Shutterstock
What is Structured Illumination Microscopy?
SIM is a fluorescence microscopy technique that is not hindered by the optical diffraction limit. SIM can be directly performed on samples that have been prepared for fluorescence microscopy with no extra preparation needed, making it a very convenient technique.
OMX 3D Structured Illumination Microscopy (3D-SIM) acquisition
A 3D Structured Illumination Microscopy (3D-SIM) acquisition from the Applied Precision OMX microscope. In this keratinocyte cell keratin filaments labelled with GFP can been seen in a structure surrounding the nucleus.
Origins of Structured Illumination Microscopy
SIM was developed to improve the resolution of fluorescence microscopy. Its development was founded on many concepts and was the joint work of many researchers in the past several decades.
Fluorescence can lead to the distortion of an image, so research to improve the resolution of the images was performed. Standing wave microscopy increased the axial resolution using axial sinusoidal illumination patterns generated by interfering opposing beams from the same laser. This approach could not be used for high-resolution imaging because it left gaps in the spatial frequencies. This was fixed by generating a short-range axial interference pattern using spatial and temporal incoherent illumination, providing in a resolution of ~100 nm. Further research discovered that moving the object between two lenses can simplify the reconstruction process. The process of reconstruction was improved upon in the early 2000s, using mathematical approaches.
Microscopy: Super-Resolution: Structured Illumination Microscopy (SIM) (David Agard)
SIM Image Reconstruction
SIM imaging is performed linearly to gain many images. The fluorescence density of the sample is multiplied by the excitation intensity in each location to give the local emission intensity, which is then imaged by the microscope. The lateral light emission is described by five overlapped shifted copies of the object structure, which contains all the information leading to the resolution improvement in SIM.
The overlapping components contribute differently to each recorded image which avoids entanglement of the information. Thus, the components are differently modulated and can be further disentangled by applying an inverse matrix. During the image reconstruction process, the components are joined in Fourier space where they overlap using a weighted average approach. Only one grating direction is used, so only a single-directional resolution gain is achieved. Consequently, the series must be acquired several times with differently orientated gratings to achieve the desired image.
Applications of Structured Illumination Microscopy in Research
SIM can be applied in many ways, giving different techniques such as 2D-SIM, 3D-SIM, total internal reflection fluorescence SIM (TIRF-SIM), etc.
Applications of Structured Illumination Microscopy to Cell Biology
Commercially available SIM devices are capable of 3D imaging up to four color channels, and they are widely used as replacements or extensions of other microscopy techniques. These methods are not available for live cell imaging due to the illumination pattern and image acquisition time. Hence, most of the publications that use SIM report on the structure of fixed samples.
3D-SIM has been used for the structural analysis of chromatin distribution in the nuclei of mammalian cells. 3D-SIM was used in up to three different color channels to simultaneously visualize chromatin in mice myoblast cells. The imaging allowed them to resolve individual nuclear pore complexes in the nuclear lamina. 3D-SIM was also used to analyze the synaptonemal complex in meiotic chromosomes of maize, to discover the transient nature of the interlocks during different stages of meiosis. Analysis of human centrosomes during different stages of the cell cycle was also performed using 3D-SIM.
3D-SIM techniques helped to discover that red blood cells infected with malaria-causing parasite Plasmodium falciparum utilize exosome-like vesicles to enable communication between parasites.
3D-SIM has been used to show that Z rings assembled during bacterial cell division in Bacillus subtilis and Staphylococcus aureus are discontinuous.
There are many more applications for SIM techniques including:
- Analysis of viral structure
- Analysis of plant structure
- Live cell imaging
- Organelle imaging
The Versatility of Structured Illumination Microscopy
Structured illuminating microscopy is a rapidly growing field that offers many high-resolution imaging techniques. SIM is extremely versatile, as it can be used on samples with minimal preparation. SIM has been used to make many important discoveries in the past and will undoubtedly lead to more in the future.
More research into improving the techniques if SIM will provide better analyzation techniques using SIM and will provide higher resolution images.
Further Reading
Last Updated: Sep 6, 2018