Among the set of new psychoactive substances (NPS), synthetic cannabinoids are one of the largest and most predominant substance classes that have challenged the forensic field ever since they first appeared 10 years ago.
Over the past seven years, synthetic cannabinoids available for purchase have experienced considerable chemical modifications making immunochemical testing inappropriate. Mass spectrometric techniques have become the gold standard for detecting and identifying non-biological and biological samples. However, due to repeated chemical modifications to the product portfolio leading to many new compounds, emerging analytical techniques have to be adapted frequently.
Providing a non-invasive sample collection with a comparatively broad window of detection, urine analysis is generally the preferred method for abstinence control. However, identification of metabolite is necessary for urine analysis because a majority of synthetic cannabinoids are metabolized extensively before renal excretion. The main phase I metabolites represent appropriate target analytes after conjugate cleavage with β-glucuronidase.
Therefore, before updating analytical techniques, it is necessary to understand the metabolism of new synthetic cannabinoids. In instances where no authentic human sample material with confirmed uptake of the specific compound is available, pooled human liver microsomes (pHLM) provide a rapid and low-cost alternative to obtain preliminary data on phase I metabolites that may be pertinent for the analysis of human urine samples.
For proof of concept of the illustrated workflow, the highly potent synthetic cannabinoid MDMB-CHMICA (methyl N-{[1-(cyclohexylmethyl)-1H-in - dol-3-yl]carbonyl}-3-methylvalinate) was selected as a model compound, since it is responsible for several intoxications all over the world and also one of the most common synthetic cannabinoids in Germany.
Table 1. HPLC conditions used in the TargetScreener workflow
| LC-MS settings |
| Mass spectrometer |
impact II
Full scan with Auto MS/MS mode, 50-600 m/z @ 4 Hz
Full scan with bbCID mode, 50-600 m/z @ 2 Hz
ESI, Positive ion mode |
| UHPLC |
UltiMate 3000RS HPLC |
| Column |
Kinetex C18 2.1x100 mm, 2.6 μm |
| Eluents |
A: 1% ACN + 0.1% HCOOH + 2 mM NH4+COO-
B: ACN + 0.1% HCOOH + 2 mM NH4+COO- |
| Gradient |
14 min gradient elution, 20 min total runtime |
| Total flow |
0.5 mL/min |
| Oven |
40 °C |
| Injection vol. |
2 μL |
Method and Material
The pHLM assay was conducted based on a standard protocol. LC-QTOF-MS analysis was carried out on an UltiMate™ 3000 RSLC connected to an impact II instrument in positive ESI mode using bbCID and data-dependent MS/MS fragmentation. LC-MS/MS datasets of the incubations sampled at time points of zero and one hour were analyzed using Mass-MetaSite™ software (Molecular Discovery).
Expected phase I biotransformations were: Di- and monohydroxylation, ester hydrolysis, dihydrodiol formation, amide hydrolysis, carboxylation, reduction, and potential combinations of these reactions. These proposed biotransformations adhere to known metabolism patterns of associated synthetic cannabinoids. An excerpt of the generated mass list offering possible metabolites is shown in Figure 1. Moreover, using MetabolitePredict software, a precursor mass list was generated for targeted MS/MS experiments (see Figure 2).
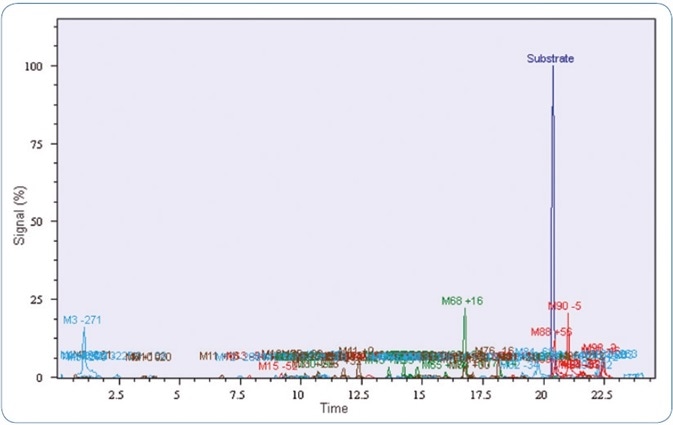
Figure 1. Overview of extracted ion chromatograms of possible metabolites in Mass-MetaSite software.
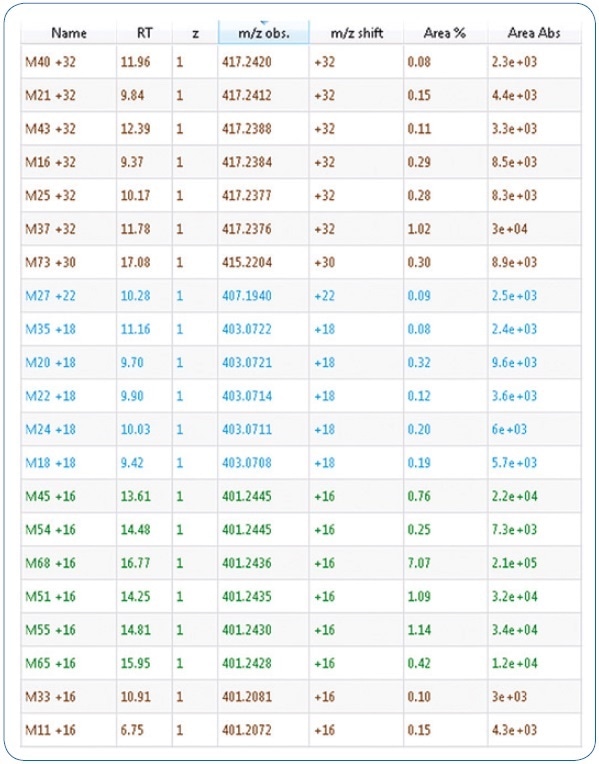
Figure 2. Excerpt of a mass list giving possible metabolites. Mass differences stated result from the respective metabolic changes of the parent compound MDMB-CHMICA.
With the help of TASQ™ software, a screening technique was created using precursor and fragment information of the known metabolites.
Results and Discussion
As mentioned above, the prevalent and highly potent synthetic cannabinoid MDMB-CHMICA was selected as a model compound. Analysis of the pHLM incubations of MDMB-CHMICA with Mass-MetaSite™ software showed 10 metabolites with at least two fragment masses each. The extracted ion chromatograms of possible metabolites in Mass-MetaSite™ software are demonstrated in Figure 3.
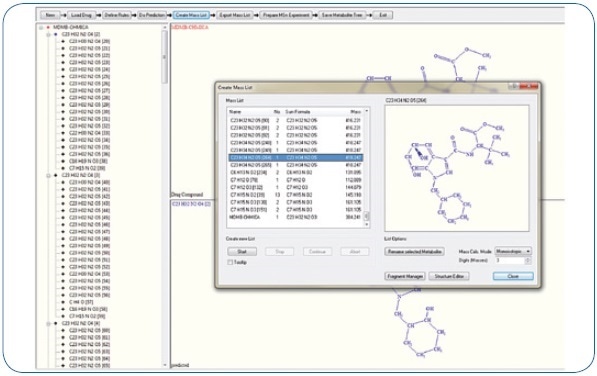
Figure 3. For MSn experiments a scheduled precursor list was generated with Metabolite Predict out of the Metabolite tools software.
The major in vitro phase I metabolites described earlier in various studies [1,2] were identified with the software. A screening technique was created in Bruker TASQ™ software using the fragment and metabolite information. The retention times were also adapted and added to the database. Using the improved TASQ™ technique, authentic forensic case samples were screened and processed (see Figures 4 and 5). In spite of varying relative abundances of the identified metabolites, the in vivo and in vitro data demonstrated excellent agreement with regard to the selected MDMB-CHMICA metabolites.
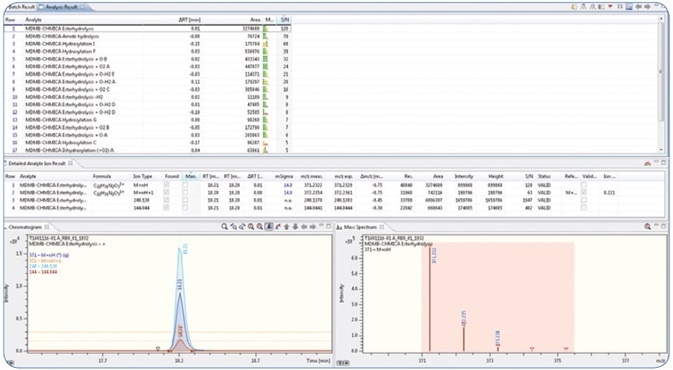
Figure 4. The identified metabolites were used together with their fragment information (qualifiers) and retention time to set up a screening method in TASQ software. Below the screening results of a urine sample positive for MDMB-CHMICA are shown.

Figure 5. Example of a pdf report generated in TASQ SW. These reports can be obtained per batch or for each sample individually.
Conclusion
- The described method can be useful for updating screening techniques with metabolite information
- The described workflow and the Mass-MetaSite™ software have been shown to be an appropriate, less time-consuming, and less laborious procedure when compared to manual data assessment
- Identification of metabolites together with parent compounds can serve as a plausibility check and may aid in estimating the time of the last uptake of drugs
- The inclusion of metabolites in screening techniques is essential when dealing with extensively metabolized analytes, for example, synthetic cannabinoids
References
1. Grigoryev et al. Forensic Toxicol. (2016) 34:316-328 doi: 10.1007/s11419-016-0319-8
2. Franz et al. Drug Test Anal. 2016 Aug 9. doi: 10.1002/a.2049. [Epub ahead of print]
About Bruker Life Sciences Mass Spectrometry

Discover new ways to apply mass spectrometry to today’s most pressing analytical challenges. Innovations such as Trapped Ion Mobility (TIMS), smartbeam and scanning lasers for MALDI-MS Imaging that deliver true pixel fidelity, and eXtreme Resolution FTMS (XR) technology capable to reveal Isotopic Fine Structure (IFS) signatures are pushing scientific exploration to new heights. Bruker's mass spectrometry solutions enable scientists to make breakthrough discoveries and gain deeper insights.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.