A team of researchers has come out with the first detailed map of the transcriptome and epitranscriptome of the virulent SARS-CoV-2 that causes COVID-19 disease, the illness that has become a pandemic ravaging both developed and less developed countries of the world. The study, published in the journal Cell in April 2020, may assist in the development of new vaccines and therapeutics.
The virus
The SARS-CoV-2 virus is a betacoronavirus, like the SARS and MERS coronavirus. The structure of the former and the SARS virus is 80% similar. While the coronaviruses were considered to be viruses affecting birds and mammals, the SARS, MERS, and COVID-19 epidemics have shown that this virus family can jump species barriers to cause human infection, and to spread between humans with ease.
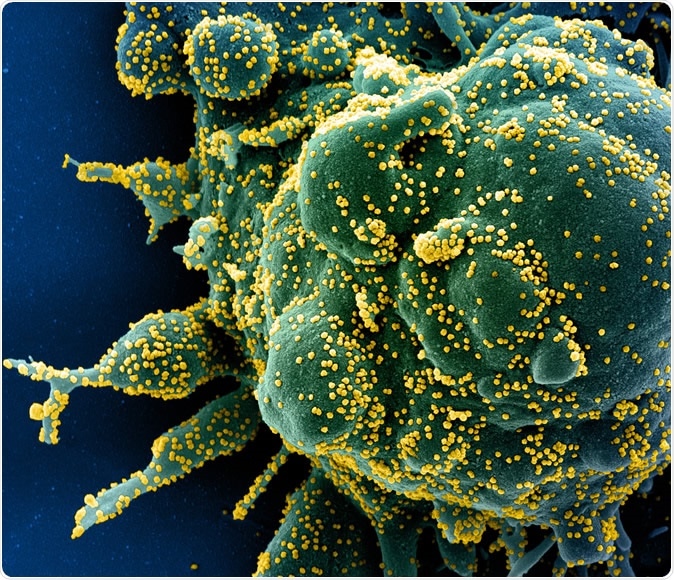
Novel Coronavirus SARS-CoV-2 Colorized scanning electron micrograph of an apoptotic cell (green) heavily infected with SARS-COV-2 virus particles (yellow), isolated from a patient sample. Image captured at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Credit: NIAID
The novel coronavirus carries a vast genome in the form of a long molecule of RNA. This is the largest genome among the families of RNA viruses, with 26-32 kilobases.
When the virus enters and infects a host cell, it replicates the RNA genome, but also generates smaller fragments of RNA called subgenomic RNAs. These are responsible for the synthesis of other viral proteins in the envelope, the spike antigen, and others that are assembled to produce complete viral particles. These could be suitable targets for producing vaccines that could interfere with the infection.
Though the genome has been sequenced, the position of the various genes has not yet been precisely defined, hindering further work. This motivated the current study, led by researchers Kim V. Narry and Chang Hyeshik, from the Center for RNA Research at the Institute for Basic Science (IBS), South Korea. Working together with the Korea National Institute of Health (KNIH) within the Korea Centers for Disease Control & Prevention (KCDC), they have successfully mapped the various gene loci within the viral genome. Experiments confirmed each of the predicted subgenomic RNAs.
How did they map the viral genome?
The team of researchers used two techniques to sequence the RNAs - DNA nanoball sequencing and nanopore direct RNA sequencing (nanopore DRS). The first is capable of accurately sequencing short fragments, at high speed, yielding a large number of such reads. The latter allows the direct sequencing of the entire RNA molecule, in contrast to the conventional procedures that require the RNA to be first cut into pieces and then reverse-transcribed to DNA before a readout can be done. Though nanopore DRS is less accurate, the long-read sequencing allows long nested transcripts to be read. It also provides data on chemical modifications directly because of its detection of RNA rather than cDNA. The two methods pair beautifully to result in a complementary analysis of the viral RNAs.
What did the study show?
The detailed map shows the entire transcriptome (viral genes) as well as the epitranscriptome (the chemical modifications on the RNA strand that do not affect the base sequence).
Earlier, researchers thought that the viral particle contained ten such subgenomic RNAs, but it is now known, as a result of this study, that there are only 9. The DNA nanoball sequencing technique showed that the viral transcriptome is composed of a large number of discontinuous transcription events.
They also had unexpected gains from the study, which revealed dozens of hitherto unknown subgenomic RNAs formed by the fusion and deletion of nucleotides on the RNA strand, as well as frameshift mutations, often accompanying the former. They also found several new chemical modifications. These epigenetic changes could be the reasons for the rapid alterations in the genetic make-up of the virus, they say. The chemical modifications could be the key to the virus’s resistance to host immune attack. Earlier research has shown that a host of RNA modifications regulate RNAs in both eukaryotes and viruses.
Furthermore, they postulate the presence of novel properties for these modified RNAs compared to the original fragments, despite the identical base sequence information for either. They aim to decipher the meaning of these changes, for instance, in the replication of the virus and in the resulting host immune responses, and to explore its lifecycle. The consistent presence of tail shortening, for instance, in modified RNA molecules, could mean an effect on the control of viral RNA stability.
The researchers sum up: “Our work provides a high-resolution map of SARS-CoV-2. This map will help understand how the virus replicates and how it escapes the human defense system.” “We firmly believe that our study will contribute to the development of diagnostics and therapeutics to combat the virus more effectively,” says Narry.
Journal reference:
Kim, D., Lee, J. Y., Yang, J. S., Kim, J. W., Kim, V. N., & Chang, H. (2020). The architecture of SARS-CoV-2 transcriptome. Cell. In press. DOI: 10.1016/j.cell.2020.04.011, https://www.cell.com/pb-assets/products/coronavirus/CELL_CELL-D-20-00765.pdf