Scientists have been working hard on finding effective ways to prevent or treat the viral infection that causes COVID-19 disease. The virus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is capable of rapid and extensive spread and evokes a powerful inflammatory reaction in a significant minority of patients.
Finding Passive Protection
Various approaches are underway to come up with an effective vaccine against the virus, including the use of viral antigens to elicit neutralizing antibody responses. Despite early results, questions remain about how long this type of vaccine’s protection will last.
Another avenue is the development of recombinant neutralizing antibodies, which may help treat existing infections before a vaccine is developed or available globally. These molecules can be chosen by their selectivity and may be invaluable in protecting older people or those in the healthcare field as well. Such molecules are already being tested in clinical trials, having been shown to confer passive immunity in animal models.
A third strategy is the use of convalescent plasma from recovered COVID-19 patients. Moreover, specific B cells that bind to the viral receptor-binding domain (RBD) that attaches to the host cell receptor ACE2 can be isolated, and the antibody sequences cloned. This has also given rise to a number of prospective antibodies which are undergoing human trials.
Synthetic Antibodies: An Overlooked Option
Synthetic antibody libraries are built on scaffolds that have been adapted to allow large-scale production of antibodies. This is an important consideration when the scope of the demand is global, and especially when the therapy needs to be effective and also affordable. Moreover, the use of uncommon formats like diabodies and scaffolds that lack Ig affinity, such as DARPins, may be required. Unlike monoclonal antibodies (mAbs), which can be generated only within mammalian cells, diabodies do not have naturally occurring IgG glycosylation sites and can be expressed in bacterial or yeast cells to allow large-scale, cost-effective production.
The Study: Finding Inhibitory Antibodies
The current study uses the Retained DisplayTM (ReD) protein display platform, which has a unique method of scaffold expression, namely, expressing and folding it in the bacterial cytoplasm before it is exposed to the extracellular environment by leakage through the membrane. This means that the scaffold must be very stable in order to undergo folding in a reducing environment, within the bacterial cell, while resisting the tendency to form disulfide bonds within individual domains that are essential for the majority of immunoglobulin domains.
One of these scaffolds is the lambda light chain family (IGLV), which has been found to contain fully-human germline antibody genes. These produce a high yield with the most commonly used heavy chain variable domain IGHV-23 after they express and fold productively. The resulting antibodies are selected at ultrafast speeds based on FACS-mediated selection of multiple target binding parameters when ReD is linked to a bacteriophage system that has high avidity - strong binding between the antibody and the antigen.
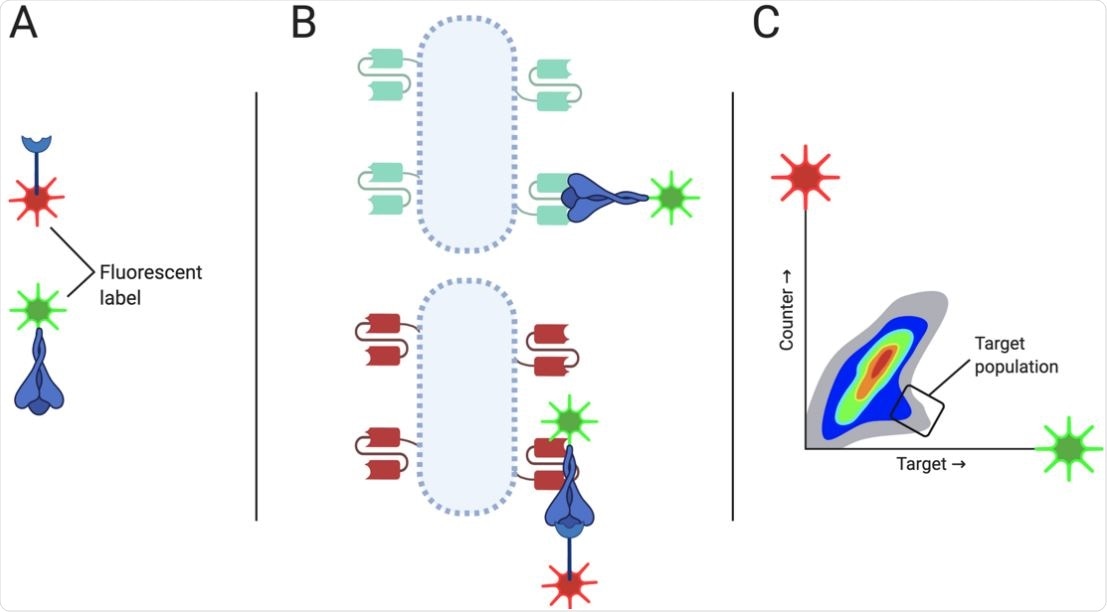
FACS strategy of screen RU167 for scFv inhibiting the SARS-CoV-2 RBD/ACE2 interaction The FACS-based screening strategy for screen RU167 to isolate antibodies that bound SARS-CoV-2 RBD and specifically inhibited co-binding of RBD to the human ACE2 protein. The viral RBD and the ACE2 protein were labeled with different fluorophores (A). Binding to cells expressing scFv clones that bound RBD and blocking the ACE2-binding site (B) would be observed and gated positively for in the FACS plot for events which were RBD-dye HIGH and ACE2-dye LOW (C).

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
This generates single-chain antibodies (scFvs), which are expressed at high yield in the bacterium Escherichia coli in multiple formats, including diabodies with bivalency, bispecifics, and scFvs. Still, all capable of binding to the specific SARS-CoV-2 RBD antigen with high affinity once converted to the native antibody format like IgG1. This shows that these scaffold molecules are robust.
Further manipulation of the antibody format is possible such as fusing the diabodies to albumin or to the albumin-binding domains, which gives them a longer half-life in serum. At present, human serum albumin is produced at an industrial scale within the yeast Pichia pastoris, which can be adapted to generate neutralizing antibodies for the current or future pandemics.
When in vitro binding assays were done using the S1 ectodomain and the human ACE2 receptor, inhibition of binding occurred only when antibodies and the S1 target were present at equimolar ratios. The antibodies required to inhibit this must, therefore, have an extremely high affinity for the S1 target since the RBD on the S1 subunit has a high affinity for ACE2.
Different Formats, Complete Protection
During a virus neutralization assay, the researchers found that both diabodies and mAb formats produced complete protection of cultured cells from infection with live SARS-CoV-2 over 4 days. This was judged by the complete inhibition of a cytopathic effect on the cells in two of four wells.
The antibody concentration required for complete inhibition over this period by the most potent clone was 5.9 nM. The current study differs from others, which have also reported neutralizing activity of antibodies in that either non-replicative pseudoviruses or short periods of infection such as one hour with live replicative virus was allowed.
In real life, the period of protection required before viral clearance occurs may vary from hours to days, making the current study more useful. Moreover, the high affinity and extended period of association, with low dissociation rates, means that the S1 protein is occupied and, therefore, unable to bind to the ACE2 to enter the host cell, despite its high affinity for the receptor.
The high stability of the various formats over the whole 4-day cell protection assay period is also noteworthy, indicating that all have comparable affinities and neutralization activity. The most potent clone showed equivalent inhibitory molar concentrations for the mAb and diabody, at 5.9 nM and 14.3 nM, respectively. This allows diabody manufacture to be an option when speed and cost are important considerations.
In summary, the researchers say, “The highest affinity mAb completely neutralized live SARS-CoV-2 virus in cell culture for four days at a concentration of 6.7 nM, suggesting potential therapeutic or prophylactic use.”
Antibodies Enhancing Virus Binding
The study points out, “Furthermore, scFvs were identified that greatly increased the interaction of the viral S1 trimer with the ACE2 receptor.”
The binding inhibition assay identified scFv clones that bound the RBD, such as to enhance S1-ACE2 binding, and this is a novel finding. It could be that the antibody clones bound the RBD at a site away from the ACE2 binding site but stabilized the trimer such that it was in a configuration suitable for binding to the ACE2, such as a locked ‘up’ position. It is worth noting that these clones were found during a screen for unique antibody clones targeting the RBD, forming 37% of the total number of clones. Thus, they are likely to emerge when the spike antigen vaccine is used as well, resulting in increased viral binding rather than inhibition.
This phenomenon, called antibody-dependent enhancement (ADE) is known to occur with MERS, for instance, using the Fc receptor on the cell surface rather than the entry receptor. In the current case, however, it uses the native ACE2 receptor and may thus worsen the infection. The researchers are continuing to analyze the process structurally to understand how this is happening. They comment, “Further investigation is required to determine if this observation has implications for the choice of epitopes when designing vaccines.”

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.