Using cryogenic electron microscopy (Cryo-EM), it is possible to study the structure of biological macromolecules, in particular, high molecular weight proteins without having to crystallize them first.
SARS-CoV is a coronavirus and thus has a lipid envelope studded with numerous spike glycoproteins. Each of these has two subunits, S1 and S2. The spike protein normally forms trimeric assemblies and mediates viral binding to the human host receptor, the angiotensin-converting enzyme 2 (ACE2), followed by viral entry into the cell.
Single-particle analysis has been performed for the SARS-CoV-2, using cryo-EM. This shows a mushroom-shaped structure, with a viral fusion bridge between the virus and host cell being formed by the viral spike protein and the ectodomain of the ACE2 receptor protein, the part that projects beyond the host cell membrane.
ACE2 Presents Drug Target
Earlier studies have revealed that the Spike receptor binding domains (RBDs) of both the SARS-CoV and the SARS-CoV-2 have comparable affinities of binding for the ACE2 ectodomain at low nanomolar levels. However, compared to earlier coronaviruses, the novel SARS-CoV-2 shows greater host cell adhesion and achieves more efficient viral entry.
Researchers have studied the structure of the wildtype inactivated SARS-CoV-2 virions by cryo-electron tomography. This showed that once the viral spike subunit S2 fuses with the host cell membrane, the post-fusion S2 trimers are scattered over the surface of the virion.
Spike-ACE2 Binding Causes Rearrangement
The ectodomain of the spike protein was expressed in prefusion form in cell culture and showed the typical trimeric form, with transmission electron microscopy (TEM) showing mushroom-shaped particles as expected. The human ACE2 ectodomain was also expressed in another cell line and incubated with the spike protein.
They carried out TEM on samples of the spike-ACE2 mixture and found that the particles were of a very different shape from the classical spike trimers. The particles were mostly smaller than the non-complexed spike trimers, with significant asymmetry.
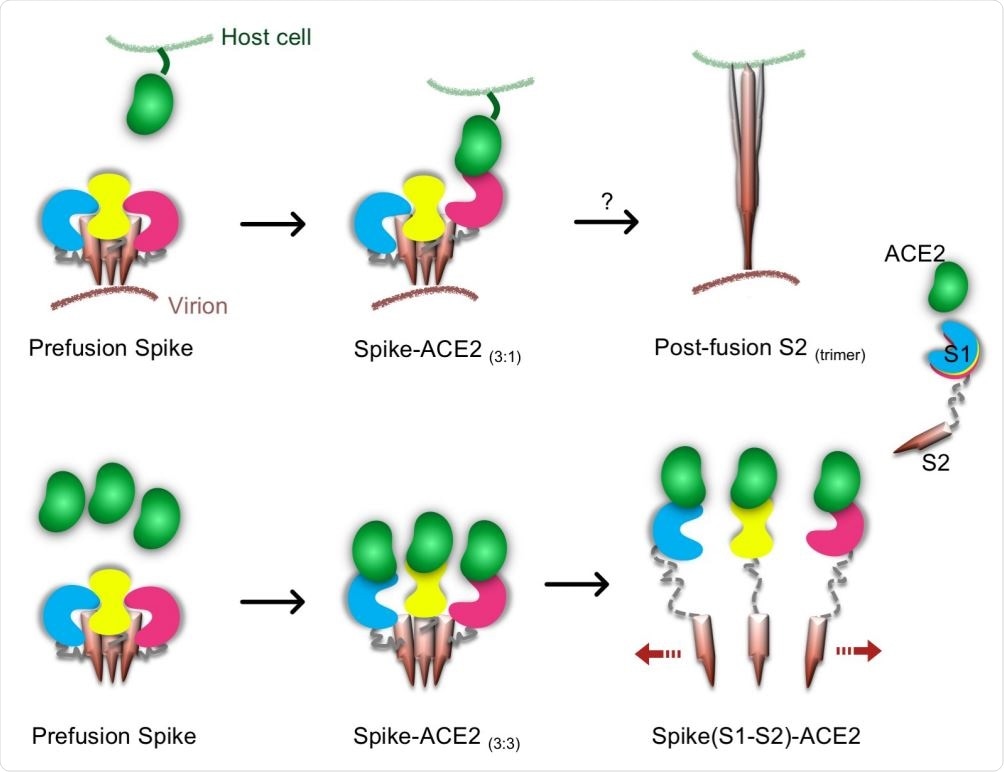
Proposed models for Spike-hACE2 complex formation and structural rearrangement. The upper row shows a possible pathway leading to a conformational change of the trimeric SARS-CoV2 Spike. In this model, one hACE2 molecule binds to one Spike S1 monomer and induces the conformational changes in the trimeric Spike. Subsequently, a post-fusion S2 trimer is formed. The lower row shows a novel proposed pathway leading to Spike trimer disassembly by hACE2. In presence of a high concentration of hACE2 molecules, a Spike-hACE2 (3:3) complex is formed. Structural clashes between the three Spike-hACE2 elements lead to their dissociation. This induces the formation of monomeric Spike-hACE2 complexes.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
The researchers concluded that the spike trimers dissociated after being incubated with the hACE2 particles for an extended period of time. This agrees with recent findings from other studies.
The variation in particle size led them to purify the complex still further, using size exclusion chromatography (SEC). They found three different particle sizes in this mixture. The first peak contained aggregated spike and hACE2, while peak 3 contained more free hACE2. Peak 2 was homogeneous and contained complexed spike-hACE2.
This was further examined by cryoEM, which showed the presence of monomeric spike protein bound to hACE2. This shows that binding to hACE2 causes the spike homotrimer to dissociate. The C-terminal domain (CTD) and N-terminal domain (NTD) of the S1 protein have undergone significant rearrangement of their structures compared to the spike monomer in the ‘up’ conformation of the spike.
This agrees with earlier reported structures of the RBD-ACE2 complex.
With a shorter incubation period, the researchers found that there was a small group of particles which corresponded to the prefusion spike trimer. The complex was shown to be made up of a mixture of spike and hACE2 in a molar ratio of 3:3, with all the RBDs being in the ‘up’ conformation and somewhat tilted from the central axis of the trimer, as earlier observed by other researchers.
Structural Comparison of Spike-hACE2 Complexes
The structural rearrangement of both the monomeric and trimeric models of the spike-hACE2 complex shows that both the CTD and NTD are shifted through ~30o ‑in order to allow the S1 protein monomer to bind the hACE2. At this point, the hACE2 and RBDs agree with earlier reported crystal structures. There are numerous steric hindrance sites at the CTD of the S1 and the neighboring region of the S2 unit in the docked model of the trimeric S1-hACE2 model.
While the exact ratio in which the spike attaches to the hACE2 for viral entry to the host cell is not yet clear, it seems that the binding of one hACE2 by one spike trimer initiates binding and initialization of viral fusion with the host cell prior to active infection. The post-fusion S2 trimer structure has also been analyzed recently, but confusion still surrounds the process by which membrane fusion is accomplished following the release of the S1-hACE2 complex.
The current study shows that a stable complex can be formed from a monomeric S1 with the hACE2, while the cryo-EM did not show any evidence of isolated S1 fragments other than in the above-mentioned equimolar Spike-hACE2 complexes. This may indicate the stability of the S1-hACE2 complex in the experimental conditions.
None of the spike-hACE2 monomeric complexes contained S2 subunits, though it was detected in the sample. The S1-S2 interface is supposed to be via a short interface loop domain harboring a furin protease cleavage site. The absence of stable trimers could increase the flexibility of this loop and thus render the S2 invisible to cryo-EM. Or else, the S2 subunits may have undergone denaturation during the preparation of the sample.
Implications
The current study shows the architecture of the spike-ACE2 protein complex. This is found to be a complex containing the spike S1 monomer and ACE2, as the result of a significant rearrangement of the structure of the isolated protein. Thus, the researchers say, binding to ACE2 causes the spike protein to change its conformation and thus undergo trimer disassembly.
This shows that ACE2 is not only a drug target to prevent but also to treat an infection with SARS-CoV-2. If such agents are developed, they could prevent the spike protein from interacting with other ACE2 molecules on other human cells, thus extending the infection.
The findings of the current study could help to understand how the S1 fragment disassembles from the complex following viral entry, which in turn could help better uncover the pathogenesis and mechanism of infection of SARS-CoV-2.
Recently, some researchers have shown that SARS-CoV-2 infection can be inhibited by a recombinant soluble hACE2 receptor, perhaps by direct competition for the host ACE2 for the viral spike protein. If sufficiently saturated with the soluble ACE2, the spike protein may not be able to bind to the host cell, they say.
This explanation becomes difficult to grasp in the light of the present findings that soluble hACE2 actually causes the trimeric spike structure to disassemble and form a stable monomeric spike-hACE2 complex. This is proposed to be via the following steps:
- Formation of a 3:3 spike-hACE2 complex
- The spike trimer becomes highly flexible
- The CTD and NTD of the spike protein undergo conformational rearrangement during the binding of the spike to hACE2
- The new S1-hACE2 complex cannot remain in trimeric form due to numerous steric obstacles and forms a monomeric complex instead by dissociation.
This hypothesis gains some support in another recent preprint that discusses the formation of a monomeric complex in response to engineered DARPin molecules.
If confirmed, this shows that soluble hACE2 may block the infection and replication of the virus as well as trigger destabilization of trimeric spike-hACE2 complexes, thus hindering the subsequent viral fusion process that is required for infection.
The researchers say, “This mechanism suggests a novel therapeutic strategy for the treatment of COVID-19, by adding soluble hACE2 to dissociate the Spike trimer of approaching viruses.”

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.