To prevent future pandemics, scientists must gather as much information about the coronavirus as possible. Previously, MERS and SARS are more deadly than severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) but have a low rate of transmission. On the other hand, SARS-CoV-2 has spread faster amongst people but has a lower risk of death than the other two viruses. Developing future coronavirus treatments will require a greater understanding of virus-host interactions.
Lei S. Qi of Stanford University, California, and colleagues published research showing a potential framework to better characterize coronaviruses' biology. While most high-resolution imaging techniques show the life cycle of coronaviruses, identifying viral or host proteins and genomes remains poorly understood. Their results use multi-color super-resolution imaging allows them to study nanoscopic parts of the coronavirus, including RNA.
The authors write:
"Broadly, we envision that future efforts will apply 3D advanced SR imaging techniques as well as the combination of single-molecule imaging and cryo-electron tomography to achieve nanometer resolution and exquisite molecular specificity simultaneously for virology research."
The study "Multi-color super-resolution imaging to study human coronavirus RNA during cellular infection" was recently published on the bioRxiv*preprint server.
How they did it
The researchers combined confocal and super-resolution microscopy to create a multi-color and multi-scale fluorescence imaging framework. The tool is designed to visualize spatial interactions between viral RNAs and different host cell areas at different stages of viral infection.
This study used the model coronavirus HCoV-229E to characterize different guide RNA structures during infection.
Results
Using the novel microscopy technique allowed the researchers to observe irregular and extended web-like clusters of guide RNA during infection. The clusters expanded as viral infection progressed with the potential to grow up to 9 square microns.
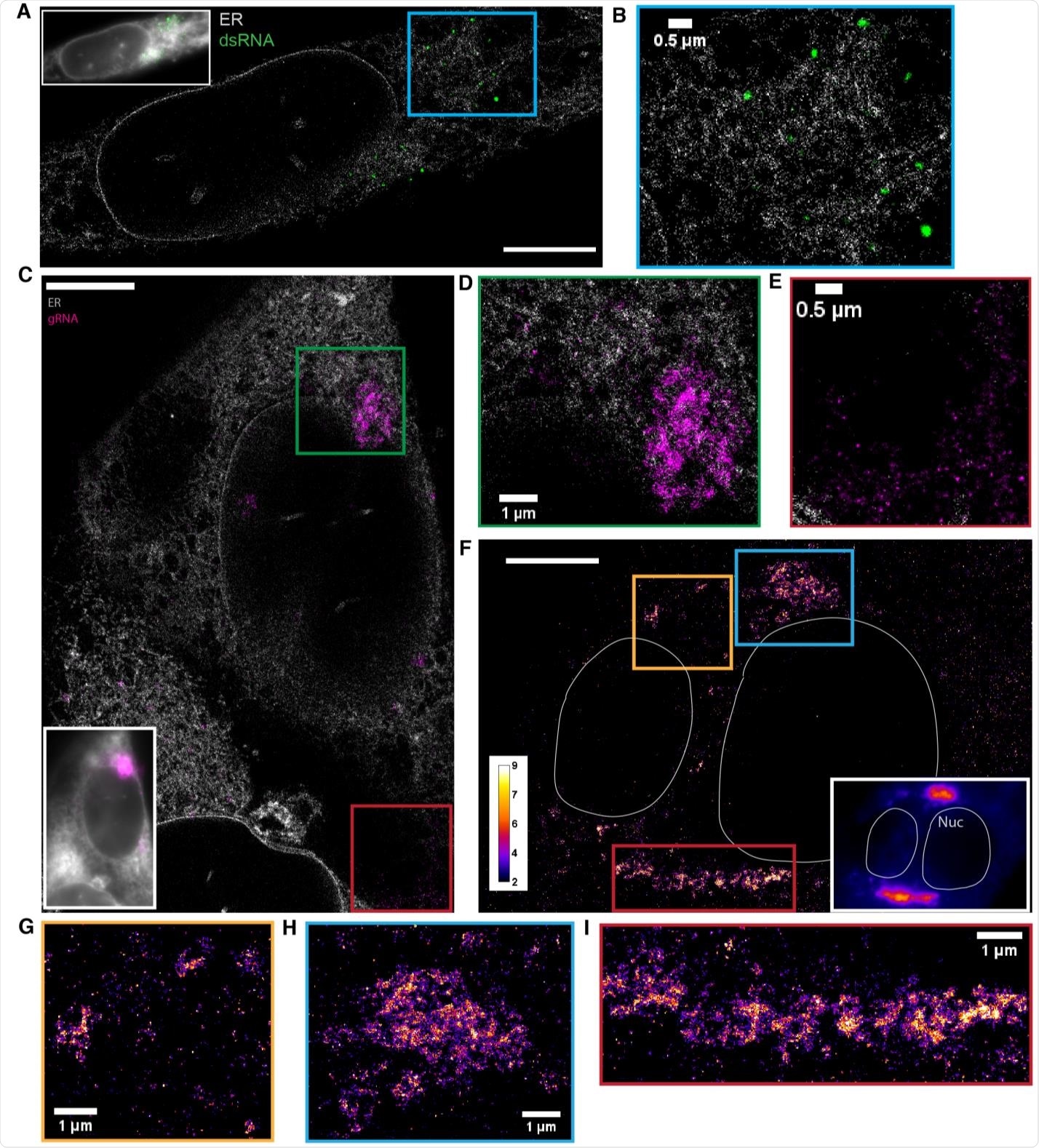
dsRNA forms puncta separated from ER while gRNA clusters frequently associate with the ER (A) Two-color SR reconstruction of a cell at 24 h p.i. where the ER (gray) and dsRNAs (green) are labeled. Scale bar: 5 μm. (B) Zoom-in of the boxed region. dsRNA forms compact puncta that appear in regions outside of the ER network. (C) Two-color SR reconstruction of a cell where the ER (gray) and gRNA (magenta) are labeled. Scale bar: 5 μm. (D) Zoom-ins of the green boxed region. gRNA forms clusters of various sizes, some of which exhibit poorly defined boundaries. The extended network of clusters is often closely associated with the ER membrane. (E) A population of the genomic RNA forms small puncta of similar sizes which are not associated with the ER (cf. Figure 3). (F) One-color SR reconstruction of gRNA at 12 h p.i. Inset: DL image of the same field of view. Scale bar: 5 µm. Color bar indicates the number of single-molecule detections within each pixel. (GI) Zoom-ins of the colored boxed regions. Large clusters of the viral genome appear perinuclear and extend up to a few microns.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The researchers observed approximately 70 nm guide RNA puncta in the cell throughout the entire infection.
"While a fraction of these puncta may be free gRNA localized away from the ER, such objects should appear larger as a result of the labeling of a 3000 bp region. Combined with the observed sparsity of these objects, we may reasonably assume that a large fraction of these nanoscale puncta are virions," wrote the team.
Indeed, the guide RNA did appear at higher densities and may indicate vesicle packets with potential virions.
The results also captured larger double-stranded RNA that formed large and circular-shaped puncta approximately 450 nm in diameter. As viral infection progressed, the number of double-stranded RNA puncta also increased. However, the size of the double-stranded RNA puncta remained constant. Based on the observations and the RNA's circular shape, the researchers suggest the puncta are likely in the lipid membranes of double-membrane vesicles.
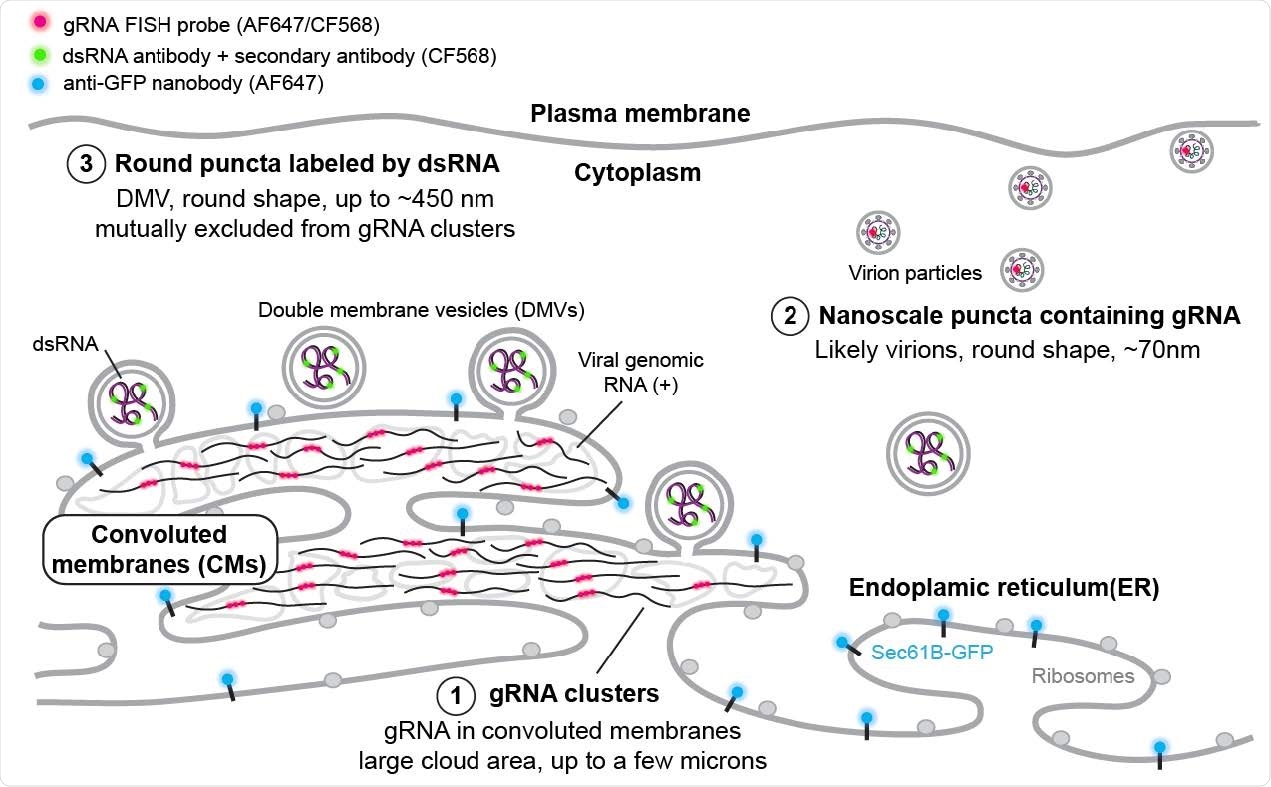
SR-based model of HCoV-229E gRNA and dsRNA distribution A model showing the spatial organization of an HCoV-229E infected cell. The ER membrane is modified to create both convoluted membranes and DMVs. Large gRNA clusters were found in association with the ER membrane, whereas packed virions that appear as well-separated nanoscale gRNA puncta are often not connected to the ER. The DMVs are filled with dsRNA to different extents. Some DMVs are still attached to the ER membrane, while others have budded off. These DMVs are separated from the convoluted membrane, where gRNA clusters appear to be the products of active replication.
The double-stranded RNA puncta did not colocalize with the endoplasmic reticulum (ER). Findings showed that puncta that are more than a hundred nanometers from the ER signal were likely cut off from the rest of the area — suggesting double-stranded RNA next to the ER could indicate double-membrane vesicles budding off the ER membrane.
"Interestingly, dsRNA puncta were not visualized by the ER membrane protein label we used in this study, suggesting that the composition and thus property of the viral manipulated membrane may be different from the rest of the ER, or the high radius of curvature of the DMVs or the membrane is too densely populated with viral proteins precluded the single-pass label native to the ER."
The nanoscale guide RNA puncta labeled virions tended to have no neighboring double-stranded RNA signal. While not colocalized, large guide RNA clusters draped with double-stranded RNA puncta at the border were noted.
When the virus was exposed to remdesivir, it decreased the confocal fluorescent signal from both guide and double-stranded RNA. Remdesivir also diminished the median side of guide RNA clusters. In contrast, the median size of double-stranded RNA puncta increased.
The findings pave the way to other avenues of research looking into how RNA is released from double-membrane vesicles for virion assembly or translation. The results confirm the usefulness of multi-color fluorescence super-resolution imaging to study these questions.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Wang J, et al. Multi-color super-resolution imaging to study human coronavirus RNA during cellular infection. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.06.09.447760, https://www.biorxiv.org/content/10.1101/2021.06.09.447760v1
- Peer reviewed and published scientific report.
Wang, Jiarui, Mengting Han, Anish R. Roy, Haifeng Wang, Leonhard Möckl, Leiping Zeng, W. E. Moerner, and Lei S. Qi. 2022. “Multi-Color Super-Resolution Imaging to Study Human Coronavirus RNA during Cellular Infection.” Cell Reports Methods 2 (2): 100170. https://doi.org/10.1016/j.crmeth.2022.100170. https://www.sciencedirect.com/science/article/pii/S2667237522000224.