Despite tremendous efforts by the global scientific community to curb the spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), by developing more efficient testing protocols, therapies, and vaccines at a rapid pace, relatively little is known about the dynamics of SARS-CoV-2 replication within cells.
Identifying the transmission and replication mechanisms of SARS-CoV-2 is vital to the development of effective drugs and vaccines and the ability to predict long-term consequences of this disease, which will aid in the development of countermeasures against the virus's evolution.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
SARS-CoV-2 entry, protein synthesis, replication, and egress
SARS-CoV-2 is an enveloped virus that enters the respiratory tract via the interaction of the receptor-binding domain on the SARS-CoV-2 spike protein and the host cell receptor, angiotensin-converting enzyme 2 (ACE-2).
The single-stranded RNA genome of the virus is then released into the host cell and is translated. Two large open reading frames - ORF1a and ORF1ab – in the genome are translated into large polyprotein complexes - pp1a and pp1ab - which are post-translationally cleaved to produce 16 non-structural proteins. The remaining ORFs in the genome encode the 4 structural proteins of SARS-CoV-2.
Upon SARS-CoV-2 infection, the virus triggers the biogenesis of replication organelles (ROs) such as double-membrane vesicles (DMVs), which protect the viral RNA from degradation by cellular RNAses during replication.
The assembly of mature viral virions occurs in the endoplasmic reticulum (ER) to Golgi intermediate compartment (ERGIC) and the egress of coronaviruses is said to happen via exocytosis.
The interactions of SARS-CoV-2 proteins with host cell proteins have only been partially studied so far, and little is known about the SARS-CoV-2 replication dynamics within the cells.
Investigating the sequence of events in the SARS-CoV-2 infection cycle by visualizing the spatiotemporal dynamics of the structural proteins
Researchers from the University of Cambridge, UK, recently investigated and characterized the sequence of events in various stages of the SARS-CoV-2 infection cycle by visualizing the spatiotemporal dynamics of the 4 structural proteins of the virus at high resolution.
They used advanced fluorescence microscopy to study the expression kinetics and spatial arrangement of the structural proteins and their interactions with host cell compartments. Their study is published on the bioRxiv* preprint server.
The researchers observed that SARS-CoV-2 structural protein expression is tightly staged, with marked differences between the nucleoprotein and the 3 transmembrane proteins. The nucleoprotein is first expressed and accumulates in convoluted 3D layers around ER membranes linked to the SARS-CoV-2 RNA replication foci.
With the help of volumetric imaging, the authors confirmed that at least one dsRNA focus is usually linked to the outer layer of the N protein compartment, which has many fused sections.
Findings offer insights into the spatiotemporal regulation of SARS-CoV-2 assembly and refines current understanding of viral replication
The results show that the N protein plays a dual role - the unmodified N protein forms a structured oligomer that is suitable for nucleocapsid assembly, and the phosphorylated N protein forms a liquid-like compartment for processing of viral genome.
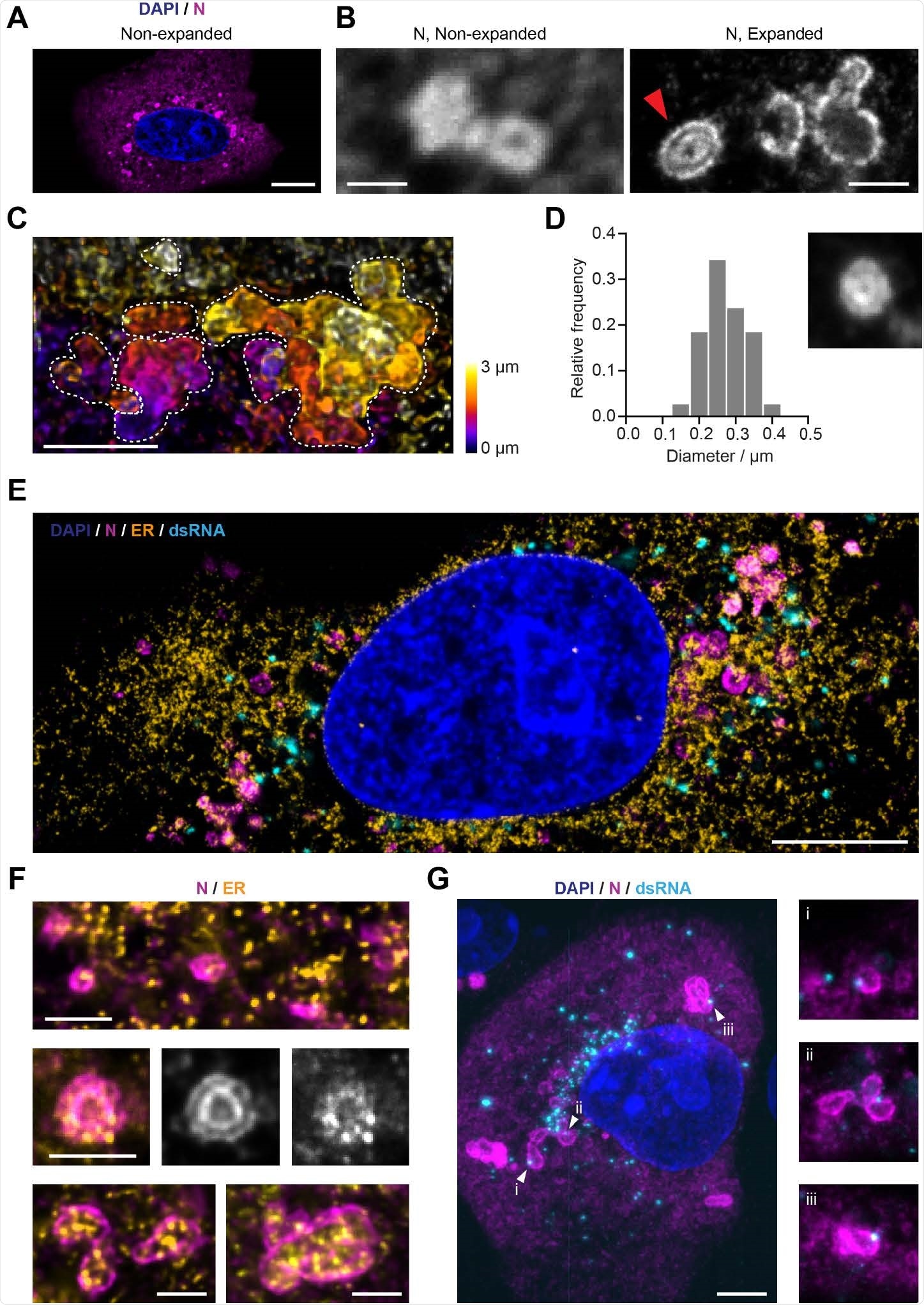
The SARS-CoV-2 nucleocapsid (N) protein is organized in layered structures that host RNA replication foci and are strongly interwoven with the topology of the endoplasmic reticulum. A) Confocal microscopy shows that the nucleocapsid protein forms punctate patterns in the cytosol of infected Vero cells (Blue: nuclei; magenta: nucleocapsid protein). Scale bar 10 μm. The numbers of puncta per cell and size increase in a stage-dependent manner (Supporting Figure 6). B) A combination of expansion and confocal microscopy shows that some of the larger nucleocapsid protein structures observed at later time points (from 10 hpi on) consist of double layers (red arrow). Scale bars 1 μm (taking into account a linear expansion factor of 4.2). C) A combination of expansion and light-sheet microscopy reveals convoluted nucleocapsid protein structures, here shown as maximum intensity projections. Each dotted line outlines the boundaries of each convoluted nucleocapsid protein structure. Scale bar 2 μm (taking into account an expansion factor of 4.2). D) Size distribution analysis of the nucleocapsid protein double-layer compartments recorded 12 hours post-infection. The inner circular layer has an average diameter of 275 nm. 38 compartments from 10 cells were analyzed. The size of the micrograph is 1.25 μm per side. E) Representative image of an expanded Vero cell stained for the nucleus (blue), ER (calnexin protein, orange), nucleocapsid protein (magenta) and dsRNA (cyan), imaged on a confocal microscope. Scale bar 5 μm (taking into account an expansion factor of 4.2). F) Details of the nucleocapsid protein (magenta) and the ER (yellow), showing that the nucleocapsid protein forms layers around ER membranes. This indicates that the nucleocapsid protein structures might be localized at double-membrane vesicles (DMVs) or packets of DMVs. Scale bars 1 μm (taking into account an expansion factor of 4.2). G) A combination of expansion and light-sheet microscopy reveals the interaction between dsRNA (cyan) and the nucleocapsid protein N (magenta). This shows that dsRNA foci sit in the layers of the N compartments. Scale bar 5 μm (taking into account an expansion factor of 4.2). Size of the smaller micrographs is 5 μm per side.
The researchers found that of the 3 transmembrane proteins, the membrane protein reaches the Golgi apparatus/ERGIC before the envelope and spike proteins.
The lysosome marker, LAMP1 relocates towards the assembly compartment and was detected in transport vesicles of SARS-CoV-2 proteins, which confirms the key role of lysosomes in SARS-CoV-2 egress.
Although a transition from a compact to a fragmented Golgi apparatus was also detected, which indicates a spike in viral egress, the reason behind this fragmentation of the Golgi apparatus is not apparent.
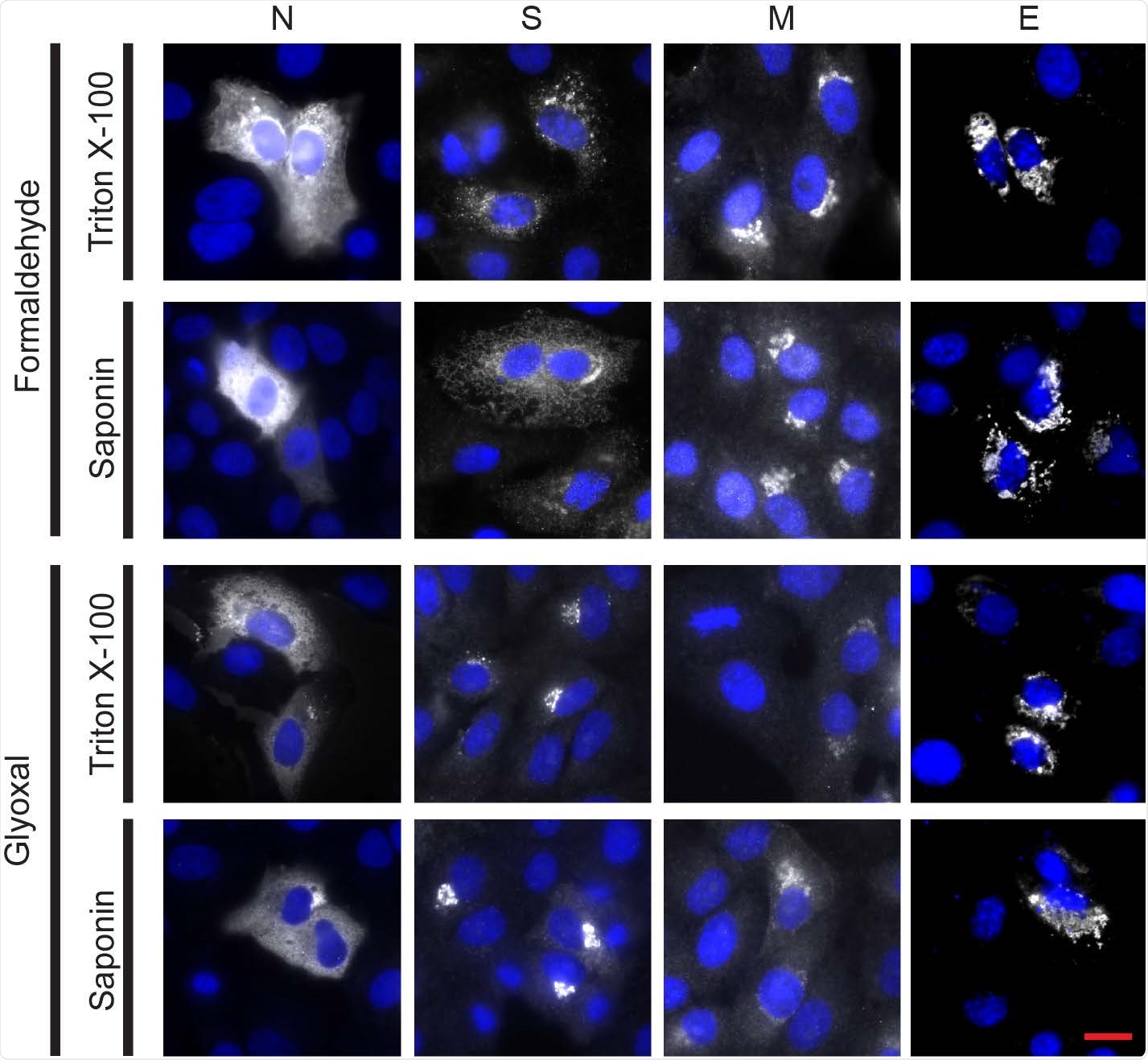
Immunostaining of the four structural proteins of SARS-CoV-2 (nucleocapsid (N), spike (S), membrane (M) and envelope (E)) was successful for all fixation conditions and selected antibodies. Vero cells were transfected to express each of the four viral proteins. The immunostaining was successful for all chemical fixation (formaldehyde or glyoxal) and permeabilization (triton X-100 or saponin) conditions tested. The spike protein pattern was different in the two fixation conditions tested, although this difference was not noticed in the infected samples. Nucleocapsid protein was detected using an Alexa Fluor 568-conjugated secondary antibody; spike, membrane and envelope proteins were detected using Alexa Fluor 488-conjugated secondary antibodies. White channel: SARS-CoV-2 proteins; blue channel: DAPI-stained nuclei. All pictures shown were acquired on a custom-built wide-field microscope. Scale bar: 20 μm.
The authors believe that further research is needed to analyze the factors contributing to the defective organization of the Golgi compartments.
The findings of this work offer new insights into the spatiotemporal regulation of SARS-CoV-2 assembly and refines the current understanding of replication of SARS-CoV-2.
"We envisage that the methods presented in this study could furthermore be used for studying the role of the non-structural proteins of SARS-CoV-2, the kinetics of the viral genome replication as well as the relationship between the viral RNA, the N protein and the viral replication organelles."
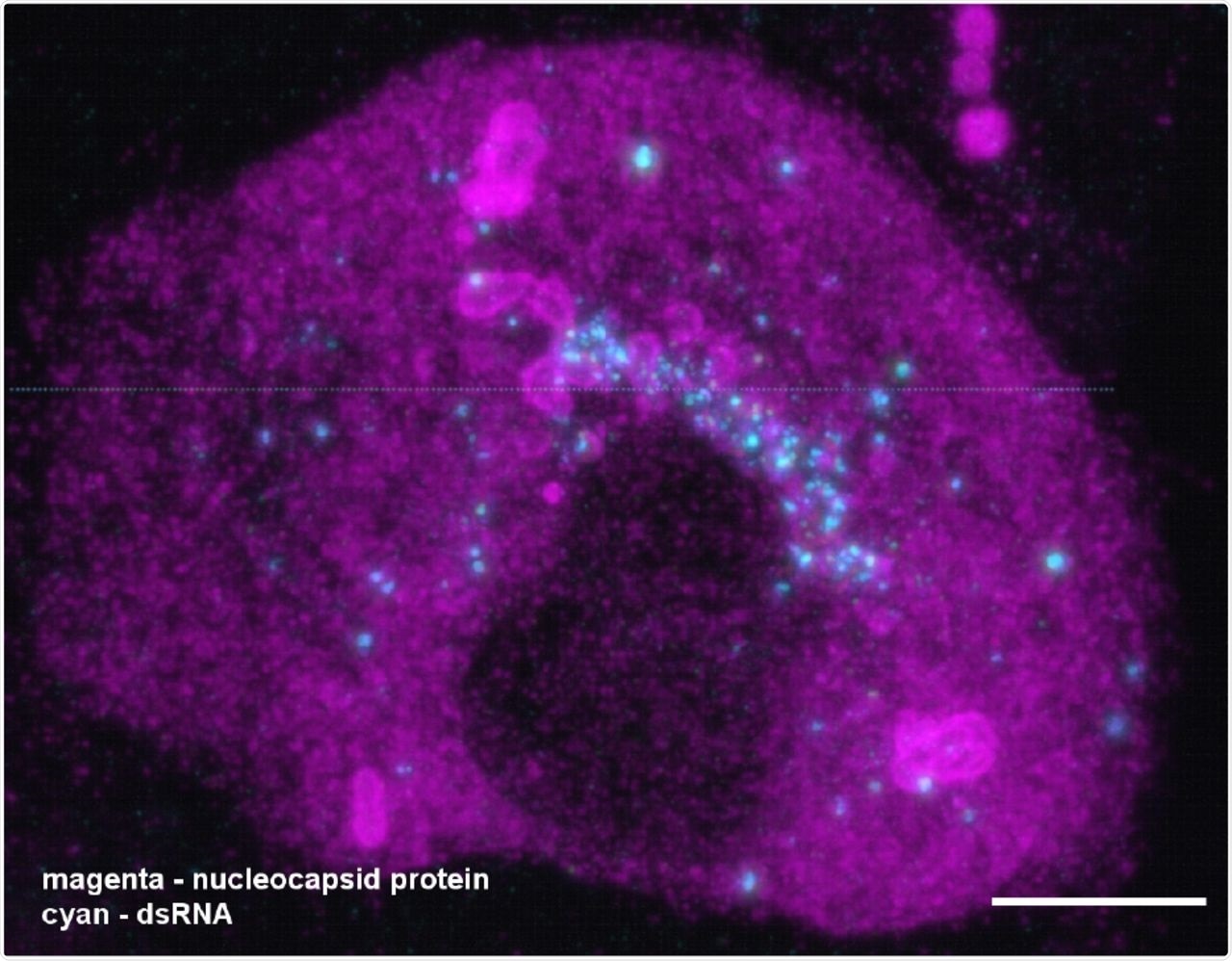
A combination of expansion and light-sheet microscopy reveals the interaction between dsRNA (cyan) and the nucleocapsid protein N (magenta) in SARS-CoV-2 infected Vero cells. The video corresponds to Figure 3G in the main manuscript. The majority of dsRNA foci are located in a region immediately adjacent to the nucleus. In addition, most N compartments contain dsRNA foci sitting in the layers of the compartments. Scale bar 5 μm (taking into account an expansion factor of 4.2).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
The SARS-CoV-2 nucleocapsid protein associates with the replication organelles before viral assembly at the Golgi/ERGIC and lysosome-mediated egress, Katharina M. Scherer, Luca Mascheroni, George W. Carnell, Lucia C. S. Wunderlich, Stanislaw Makarchuk, Marius Brockhoff, Ioanna Mela, Ana Fernandez-Villegas, Max Barysevich, Hazel Stewart, Maria Suau Sans, Charlotte L. George, Jacob R. Lamb, Gabriele S. Kaminski-Schierle, Jonathan L. Heeney, Clemens F. Kaminski, bioRxiv, 2021.06.15.448497; doi: https://doi.org/10.1101/2021.06.15.448497, https://www.biorxiv.org/content/10.1101/2021.06.15.448497v1
- Peer reviewed and published scientific report.
Scherer, Katharina M., Luca Mascheroni, George W. Carnell, Lucia C. S. Wunderlich, Stanislaw Makarchuk, Marius Brockhoff, Ioanna Mela, et al. 2022. “SARS-CoV-2 Nucleocapsid Protein Adheres to Replication Organelles before Viral Assembly at the Golgi/ERGIC and Lysosome-Mediated Egress.” Science Advances 8 (1). https://doi.org/10.1126/sciadv.abl4895. https://www.science.org/doi/10.1126/sciadv.abl4895.