A high prevalence of the H3 subtype avian influenza virus (AIV) has been observed among waterfowl globally. It causes asymptomatic or mildly symptomatic infection in birds. In addition, a high cross-species transmission ability makes the virus a potential source for the origination of other animal influenza viruses.
The first human case of AIV infection (H3N8) was detected in China in April 2022. The patient was present with recurrent fever and severe pneumonia. The second case with mild influenza symptoms was detected in May 2022. These cases raised global concern about the possibility of a significant public health threat by H3N8 AIVs.
In China, a high circulation of H3 AIVs has been observed in poultry and wild birds. Many studies have reported combinations of H3 with multiple neuraminidase (NA) subtypes, especially H3N2 and H3N8. These viruses belong to the Eurasian lineage, which is highly prevalent among wild birds in Eurasia. Genetic exchange events (reassortments) between different AIVs mainly occur in live poultry markets.
In the current study, scientists have conducted country-level AIV surveillance in poultry-associated environments during 2009 – 2022 and performed a large-scale genetic analysis to provide a comprehensive overview of the evolution of H3 AIVs in China.
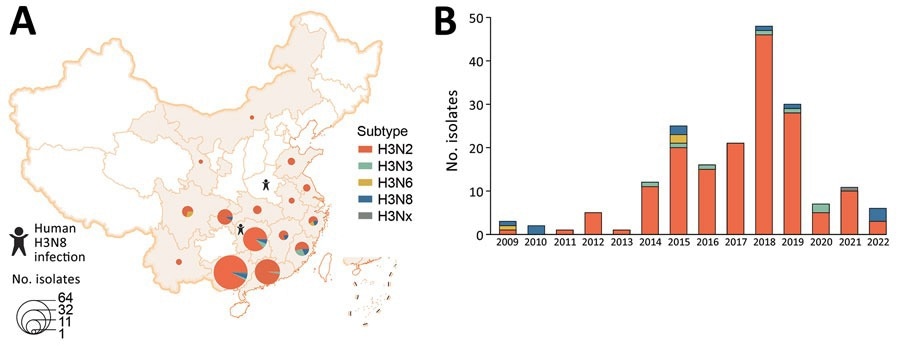
Spatial and temporal distribution of avian influenza virus subtype H3 isolated from poultry-associated environments, China, 2009–2022. A) Spatial distribution of environmental H3 subtype viruses. One H3 isolate without neuraminidase (NA) subtype was designated as H3Nx. Provinces where human infections with H3N8 were reported are noted. B) Number of environmental H3 subtype isolates per year. This figure includes all H3 isolates sequenced by the Chinese National Influenza Center.
Study design
During the study period, the scientists collected samples from poultry-related environments across 31 provinces in China. They followed the AIV surveillance guideline of the Chinese Center for Disease Control and Prevention for sample collection.
From these samples, they isolated and sequenced 188 H3 AIVs and deposited the sequences in the GISAID EpiFlu database. By aligning the sequences with available sequences from the GISAID EpiFlu database, they reconstructed maximum-likelihood phylogenies of all viral segments.
They further classified the phylogenetic trees into divergent lineages or sublineages. Furthermore, they estimated the time to the most recent common ancestor (tMRCA) of human H3N8 viruses.
Important observations
The large-scale sequencing analysis conducted in the study identified four sublineages (China-1, China-2, China-3, and China-4) of H3 AIVs in domestic ducks in China. Hemagglutinin (HA) genes of these sublineages were evolved from the Eurasian lineage. These viruses were established in China poultry via multiple introductions from wild birds from Eurasia.
The surveillance data revealed that H3 AIVs belonging to China-1 and China-2 sublineages are currently co-circulating in poultry. Among these sublineages, China-1 is predominating. In addition, the data revealed that H3N2 predominated among H3 viruses in poultry-related environments during 2009 – 2022.
The phylogenetic analysis revealed an intense genetic exchange between H3 AIVs, leading to the generation of multiple genotypes. A total of 126 genotypes were identified from 284 H3 AIVs during 2009 – 2022. Further analysis revealed that 73 genotypes (G1–G73) emerged from 212 H3N2 genomes, 11 genotypes (G1–G11) from 14 H3N3, 17 genotypes (G1–G17) from 25 H3N6, and 25 genotypes (G1–G25) from 33 H3N8.
Although most of the genotypes were transient, H3N2 G23 appeared to have stabilized recently and was predominantly circulating in Southern China. H3N8 G25, which showed cross-species transmission from binds to humans, was found to acquire HA genes from the China-1 sublineage, NA genes from the North American N8 lineage, and all 6 internal genes from poultry H9N2 ZJ-HJ/07 sublineage viruses.
The estimation of tMRCA of all 8 segments revealed that the H3N8 G25 viruses were generated by reassortments between H3N2 G23, wild bird H3N8, and poultry H9N2 before February 2021. Furthermore, the findings revealed that H3N8 G25 viruses acquire human-adapted mutations, such as 228G/S in the HA gene and E627K/V in the PB2 gene, after infecting humans. These observations highlight the potential of newly emerged H3N8 AIVs in creating future pandemics.
Study significance
The study provides a detailed overview of the evolution of H3 AIVs in China. According to the study findings, H3N8 G25 viruses, which have an increased ability to bind human receptors and low population immunity, have raised global concern over the possibility of future pandemics.
Since mammal-adapted and drug-resistance mutations rarely occur in H3 AIVs, the scientists highlight the need for vaccine and drug stockpiles for the potential pandemic preparation. Moreover, they recommend continuous surveillance for H3 AIVs and risk assessment for potential pandemic preparedness.