Background
Since SARS-CoV-2 was first identified at the end of 2019, several mutant strains of this virus have emerged. Of these include SARS-CoV-2 variants of concern (VOCs), which are considered to be the most highly transmissible and virulent strains of this virus.
The SARS-CoV-2 Delta VOC (B.1.617.2), which was first identified in India in December of 2020, quickly spread worldwide and became the dominant circulating strain of SARS-CoV-2. In fact, one recent study found that SARS-CoV-2 Delta variant-infected individuals had viral loads that were more than 1,000 times higher as compared to individuals who were infected with the original SARS-CoV-2 strain, thus making this variant the most contagious strain of SARS-CoV-2 to date.
In addition to SARS-CoV-2, the influenza virus is also highly contagious and continues to pose a significant threat to public health each year. Since both of these viruses can be transmitted at high levels during the same seasons, it is essential to develop a multivalent vaccine that could simultaneously protect immunized individuals against both the coronavirus disease 2019 (COVID-19) and influenza.
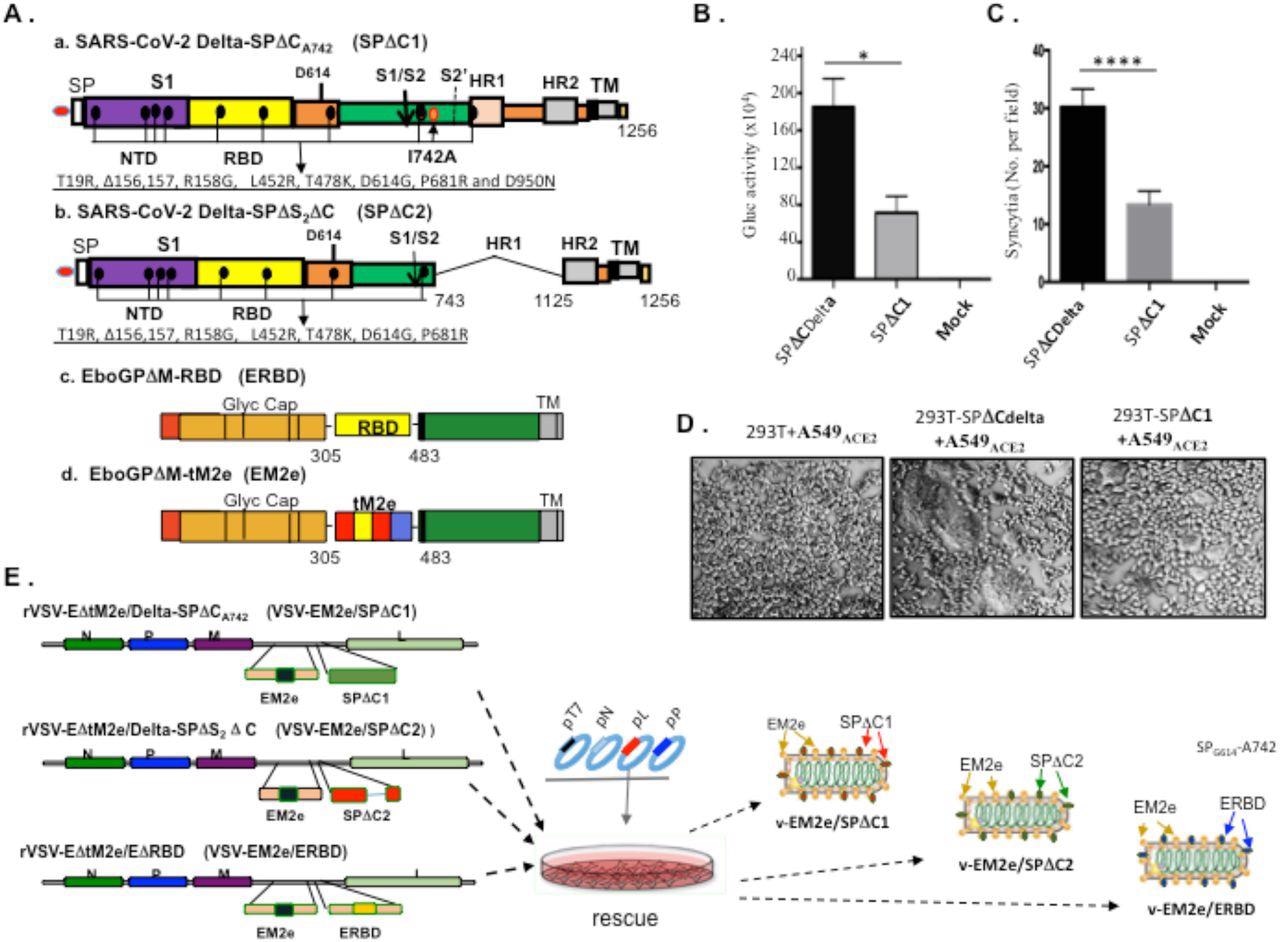 Construction and rescue of rVSV Delta SP and influenza M2e bivalent vaccines. A) Schematic diagram of the Delta SPΔC and EboGPΔM-tM2e immunogens present in the bivalent vaccines. a. SARS-CoV-2 Delta-SPΔCA742 (SPΔC1), containing a C-terminal 17 aa (DEDDSEPVLKGVKLHYT) deletion and a I742A mutation as indicated. The nine mutations in Delta SP are listed in lower part. b. Delta SPΔC2, containing the C-terminal 17 aa deletion and another 381 aa (encompassing aa744 to aa1124) deletion in S2 domain. The eight mutations in SPΔC2, are listed in lower part. c. EboGPΔM-RBD, the RBD of SARS-CoV-2 was used to replace the MLD domain in EboGP. d. EboGPΔM-tM2e, four copies of influenza virus M2 ectodomain (24 aa) polypeptide (tM2e) replaced the MLD domain in EboGP. B) The attenuated virus entry of SPΔC1. A549ACE2 cells were infected with equal amounts of SPΔCDelta-PVs or SPΔC1-PVs (adjusted by P24) carrying Gaussia luciferase (Gluc) gene, as indicated. At 48hrs after infection, the Gluc activity in the supernatant of different infected cultures was measured. Data represent Mean ±SD of two replicates from a representative experiment out of three performed. C and D) The attenuated cell-to-cell fusion ability of SPΔCDelta- or SPΔC1-mediated syncytia formation was analyzed by co-culturing the SPΔCDelta- or SPΔC1-expressing 293T cells with A549ACE2 cells. The amounts of syncytia were counted after 24 hrs in 5 different views of microscope (C), and were also imaged under bright-field microscopy (D). E) Schematic diagram of VSV-EM2e/SPΔC1, VSV-EM2e/SPΔC2 and VSV-EM2e/ERBD and the virus rescuing procedures. 293T and Vero E6 co-culture cells were co-transfected with VSV-ΔG-EM2/SPΔC1, VSV-ΔG-EM2/SPΔC or VSV-ΔG-EM2/RBD, and helping plasmids (T7, N, L, P plasmids). The supernatants containing V-EM2e/SPΔC1, V-EM2e/SPΔC2 and V-EM2e/ERBD viruses were used to infect Vero E6 cells to generate the rVSV stocks.
Construction and rescue of rVSV Delta SP and influenza M2e bivalent vaccines. A) Schematic diagram of the Delta SPΔC and EboGPΔM-tM2e immunogens present in the bivalent vaccines. a. SARS-CoV-2 Delta-SPΔCA742 (SPΔC1), containing a C-terminal 17 aa (DEDDSEPVLKGVKLHYT) deletion and a I742A mutation as indicated. The nine mutations in Delta SP are listed in lower part. b. Delta SPΔC2, containing the C-terminal 17 aa deletion and another 381 aa (encompassing aa744 to aa1124) deletion in S2 domain. The eight mutations in SPΔC2, are listed in lower part. c. EboGPΔM-RBD, the RBD of SARS-CoV-2 was used to replace the MLD domain in EboGP. d. EboGPΔM-tM2e, four copies of influenza virus M2 ectodomain (24 aa) polypeptide (tM2e) replaced the MLD domain in EboGP. B) The attenuated virus entry of SPΔC1. A549ACE2 cells were infected with equal amounts of SPΔCDelta-PVs or SPΔC1-PVs (adjusted by P24) carrying Gaussia luciferase (Gluc) gene, as indicated. At 48hrs after infection, the Gluc activity in the supernatant of different infected cultures was measured. Data represent Mean ±SD of two replicates from a representative experiment out of three performed. C and D) The attenuated cell-to-cell fusion ability of SPΔCDelta- or SPΔC1-mediated syncytia formation was analyzed by co-culturing the SPΔCDelta- or SPΔC1-expressing 293T cells with A549ACE2 cells. The amounts of syncytia were counted after 24 hrs in 5 different views of microscope (C), and were also imaged under bright-field microscopy (D). E) Schematic diagram of VSV-EM2e/SPΔC1, VSV-EM2e/SPΔC2 and VSV-EM2e/ERBD and the virus rescuing procedures. 293T and Vero E6 co-culture cells were co-transfected with VSV-ΔG-EM2/SPΔC1, VSV-ΔG-EM2/SPΔC or VSV-ΔG-EM2/RBD, and helping plasmids (T7, N, L, P plasmids). The supernatants containing V-EM2e/SPΔC1, V-EM2e/SPΔC2 and V-EM2e/ERBD viruses were used to infect Vero E6 cells to generate the rVSV stocks.
About the study
In the current study, the researchers generated three attenuated replicating rVSV vaccine candidates. These rVSV-based vaccines co-express the SARS-CoV-2 Delta variant spike protein (SP) or the receptor-binding domain (RBD), as well as four copies of the highly conserved M2 ectodomain (M2e) of influenza A that have been fused with the DC-targeting/activation domain derived from the Ebola virus glycoprotein (EBOV GP).
The researchers utilized the VSV-based vaccine platform as a result of its ability to induce rapid and robust immunity against viral antigens following a single immunization. Furthermore, previous VSV vaccine candidates have successfully and simultaneously protected immunized individuals against several pathogens including the Ebola, Zika, Nipah, and human immunodeficiency viruses.
Study findings
The bivalent rVSV vaccines constructed in the current study simultaneously express an EboGPΔM-M2e fusion protein (EM2e) and an EboGPΔM-SARS-CoV2 RBD fusion protein (ERBD), a full-length SPΔCDelta (SPΔC1), or an S2-deleted SPΔCDelta (SPΔC2). Taken together, a total of four different vaccine candidates were assessed in this study.
To investigate the safety profile of the vaccine candidates, the researchers conducted replication kinetics of rVSV-based vaccines in various cell lines and compared these results to cells treated with rVSV expressing VSV-G. These experiments demonstrated that rVSV-based vaccines do not target CD4+ T-cells, which is important due to their role in protecting the immune system from attack. Furthermore, the rVSV vaccine candidates were found to have reduced replication ability during these in vitro experiments, thus demonstrating their negligible cytopathic effect in cells.
To evaluate the ability of these vaccine candidates to induce specific immune responses against both SARS-CoV-2 and influenza viruses, the researchers immunized Balb/c mice with each bivalent VSV vaccine intramuscularly (IM) or intranasally (IN) at Day 0 and Day 14.
Replication kinetics studies demonstrated that the replication of these rVSV viruses was significantly attenuated as compared to wild-type VSV and could not infect CD4+ T-lymphocytes, thus suggesting highly attenuated characteristics of these rVSV-based vaccines. Additionally, the immunization study and neutralizing analysis revealed that the sera from V-EM2e/SPΔC1- and V-EM2e/SPΔC2-immunized mice exhibited significantly higher neutralizing activity against SARS-CoV-2 SPDelta-PV infection than against SPWT-PV infection.
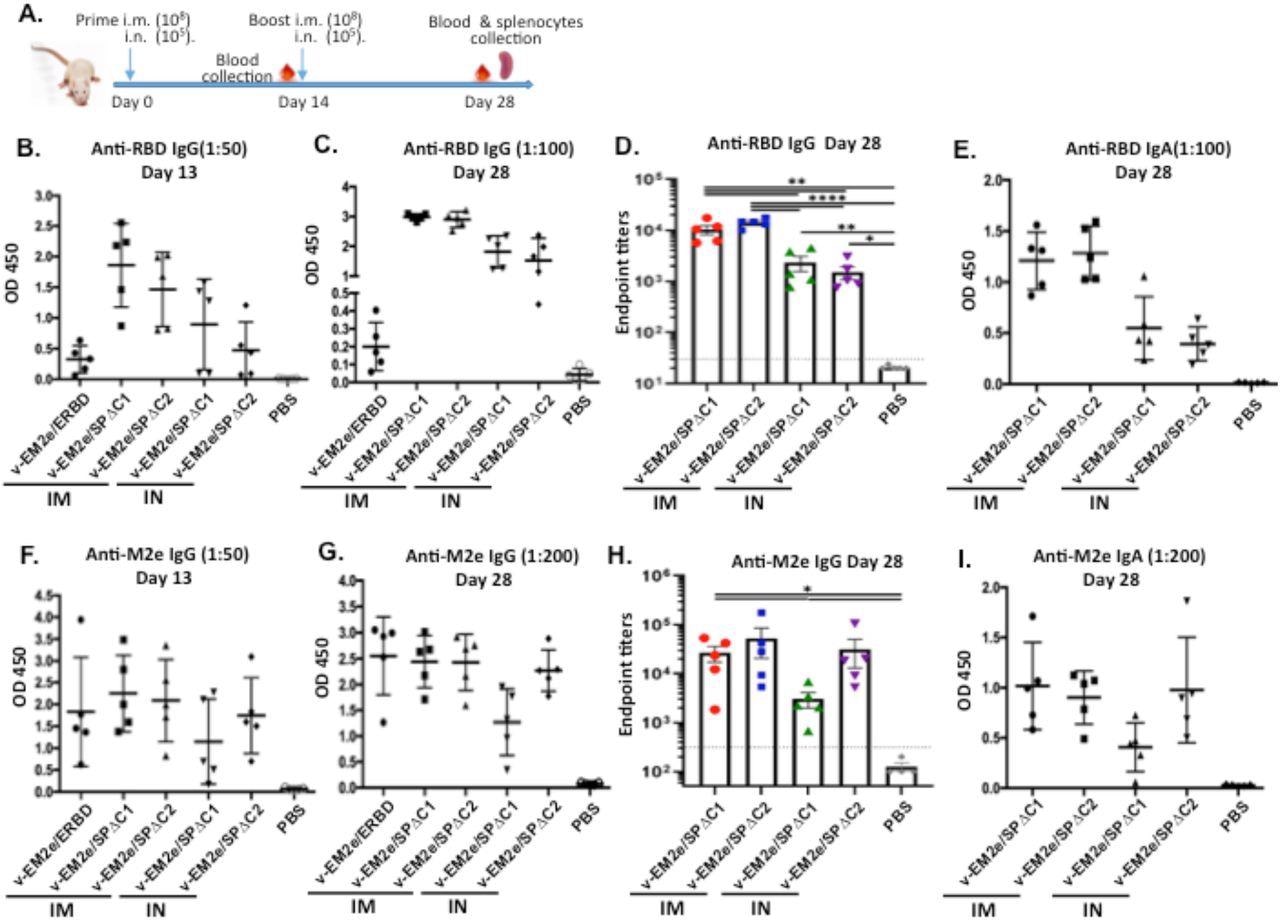 Anti-SARS-CoV-2 RBD and anti-influenza M2e immune responses induced by immunization with different bivalent VSV vaccine candidates. A) Schematic of the bivalent rVSV vaccine candidates immunization protocol in mouse. Balb/c mice were immunized with V-EM2e/SPΔC1, V-EM2e/SPΔC2 or V-EM2e/ERBD via intramuscular (IM) or intranasal (IN) routes, as indicated. The mice sera were collected at day 13 and 28 and were measured for anti-SARS-CoV-2 RBD IgG and IgA antibody levels (B-D) or measured for anti-M2e IgG and IgA antibody levels (F-H). E, D) The anti-SARS-CoV-2 RBD and anti-M2e IgA antibody levels at 28 day. Data represent Mean ±SD. Statistical significance was determined using unpaired T-test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Anti-SARS-CoV-2 RBD and anti-influenza M2e immune responses induced by immunization with different bivalent VSV vaccine candidates. A) Schematic of the bivalent rVSV vaccine candidates immunization protocol in mouse. Balb/c mice were immunized with V-EM2e/SPΔC1, V-EM2e/SPΔC2 or V-EM2e/ERBD via intramuscular (IM) or intranasal (IN) routes, as indicated. The mice sera were collected at day 13 and 28 and were measured for anti-SARS-CoV-2 RBD IgG and IgA antibody levels (B-D) or measured for anti-M2e IgG and IgA antibody levels (F-H). E, D) The anti-SARS-CoV-2 RBD and anti-M2e IgA antibody levels at 28 day. Data represent Mean ±SD. Statistical significance was determined using unpaired T-test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
All vaccine candidates also successfully induced a significant influenza-specific immune response. In fact, the lead vaccine candidate of V-EM2/SPΔC1 when administered either IM or IN, effectively protected mice from lethal H1N1 influenza virus infection.
The IM administration of V-EM2e/SPΔC1 or V-EM2e/SPΔC2 induced higher titers of neutralizing antibodies and more robust cellular immune responses as compared to when these vaccines were administered IN. However, the dose of the VSV vaccine used for IN administration was 1,000-fold lower than that for IM administration, thus suggesting the possible need to increase the dose of this vaccine to achieve stronger immune responses when administered IN.
The researchers observed strong T-cell responses that were characterized by high levels of secreted cytokines including interferon (IFN)-γ and interleukin (IL)-4 in splenocytes from mice stimulated with rVSV-based vaccine candidates. A significant difference in the ratios of Th1- cytokine/Th2-cytokine (IFN-γ/IL-5) was observed between the samples isolated from the IM- and IN-immunized mice. This suggests that IM immunization triggered a Th1-biased but relatively Th1/Th2-balanced cellular response, while IN immunization induced an extremely Th1-biased cellular response.
There is a strong likelihood that Th2 responses are associated with antibody-dependent enhancement (ADE) of infection in mice vaccinated with inactivated vaccines and challenged by SARS-CoV-1. However, the researchers indicate that the observed Th1-biased cellular response was likely due to both IM and IN administration of the rVSV-based SARS-CoV-2 vaccines.
Conclusions
The findings of the current study demonstrate that the rVSV-based vaccines described here, particularly the lead vaccine candidate V-EM2e/SPΔC1, can generate high levels of neutralizing antibodies against the SARS-CoV-2 Delta strain, as well as both the H1N1 and H3N2 influenza viruses. Further investigation is needed to confirm the efficacy of this vaccine candidate against both the SARS-CoV-2 Delta variant and influenza strains in higher animal models, particularly non-human primates before these vaccine candidates can be tested in humans.
“An important unique feature of the vaccine platform in this study is that it can simultaneously protect against SARS-CoV-2 and influenza viruses.”
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.