Background
Omicron is the fifth SARS-CoV-2 variant of concern (VOC) to be identified since the start of the coronavirus disease 2019 (COVID-19) pandemic. At the beginning of 2022, the Omicron sub-variant BA.1 replaced the Delta VOC, which was previously the dominant circulating strain worldwide. The Omicron BA.2 sublineage subsequently outcompeted BA.1 and has been responsible for the most recent wave of SARS-CoV-2 infections.
Only about 13% of the SARS-CoV-2 genome encodes for the S protein. Nevertheless, the Omicron S protein contains the greatest number of mutations as compared to other SARS-CoV-2 variants.
More specifically, the Omicron BA.1 and BA.2 S proteins share about 20 mutations in this region, 12 of which reside in the receptor-binding domain (RBD). Fourteen of these mutations are specific for BA.1 and nine for BA.2. The remaining lineage-specific S mutations are found in the N-terminal domain (NTD) of the S protein.
New Omicron sub-variants are continuously emerging, some of which include BA.2.12.1, BA.4, and BA.5. These mutations contain additional S mutations in amino acid (AA) residues L452 and F486 that might further increase their immune evasion properties and transmission fitness.
All current COVID-19 vaccines are based on the SARS-CoV-2 Wuhan-Hu 1 S antigen. As a result, mutations in the NTD or RBD of the Omicron S could increase the resistance of these strains to neutralizing antibodies (nAbs). These mutations may also be crucial determinants of the higher transmissibility and reduced pathogenicity of these variants.
To date, previous studies have not described the functional impact of most Omicron S mutations.
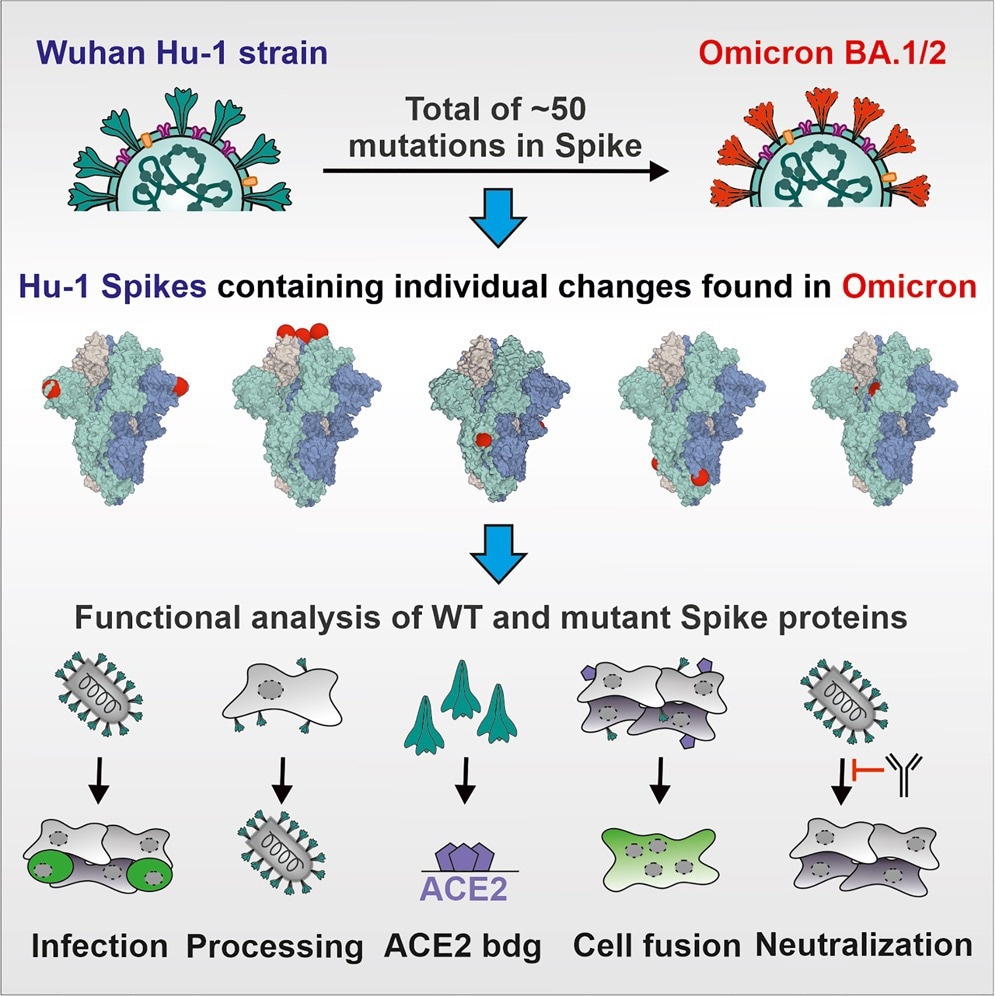
Graphical abstract
About the study
In the present study, researchers introduced 48 S mutations of Omicron sublineages BA.1 and BA.2 into the Wuhan-Hu1 S through site-directed mutagenesis. Sequence analysis of the S genes was subsequently performed to confirm that all constructs contained the desired mutations. The impact of each AA change on viral expression, proteolytic processing, infectivity, and susceptibility to nAbs and sera from vaccinated individuals was also conducted.
Vesicular stomatitis virus particles pseudotyped (VSVpp) with the parental and mutant S proteins were used to analyze the impact of Omicron BA.1 and BA.2 S mutations. S proteins lacking an artificial deletion of the C-terminal endoplasmic reticulum (ER)-retention motif were also used for in vitro assays.
The researchers also used full-length S proteins containing a simian virus 5 (V5) epitope tag for analysis of Omicron S expression and processing. Several assays were also performed to quantify the number of VSVpp infected Caco-2 cells over time.
Study findings
The BA.1 and BA.2 S proteins exhibited significantly reduced infection efficiencies as compared to the Wuhan Hu-1 S protein, while the S protein of the Delta VOC was associated with significantly increased activity. Most of the 20 AA changes in the S proteins shared between BA.1 and BA.2 did not affect the efficiency of VSVpp infection. Furthermore, most of the BA.1 and BA.2 specific NTD mutations caused minor effects on VSVpp infectivity.
Individual RBD mutations including S371F/L, S375F, and T376A strongly impaired S-containing VSVpp infectivity and S processing. More specifically, S375F almost completely disrupted S function and processing, with the adjoining BA.2-specific T376A mutation exhibiting similar disruptive effects. Although these mutations significantly reduced S-mediated infection, they had moderate effects on neutralization by vaccinated sera.
Mutations in the three serine residues of S371, S373, and S375 severely impaired the ability of the Wuhan Hu-1 S protein to mediate virus-cell and cell-cell fusion. Furthermore, the S371L, S373P, and S375F AA mutated residues promoted interprotomer interactions between the “down” RBDs.
Conversely, most lineage-specific NTD mutations such as A27S, T95I, Δ142-144, G142D, INS214EPE, L212I, and V213G, were least effective in reducing S sensitivity to neutralization by sera from vaccinated individuals. This finding supports previous reports suggesting that the NTD is a target of nAbs in vaccinated sera.
Multiple shared mutations in the RBD of BA.1 and BA.2 S proteins reduced the sensitivity of VSVpp to neutralization by vaccinated sera. In addition, N440K or G446S mutations conferred resistance to therapeutic antibodies imdevimab, whereas changes to E484A and Q493R altered the efficacy of bamlanivimab. These results suggest that single AA changes can induce resistance of the variants against nAbs.
The current study highlighted that Omicron sequences in global databases that appear to encode S residues at positions 371, 373, and 375 are erroneous. Since some apparent variations in the S proteins of emerging SARS-CoV-2 variants result from sequencing artifacts, it is important to consider these while studying mutations in particular AA sequences.
Moreover, the molecular mechanisms responsible for several mutations in the Omicron S protein remain unknown. For example, BA.2-specific changes in the NTD severely reduced S-mediated infection and processing; however, these changes do not affect known functional domains.
Similarly, a BA.2-specific N856K substitution generated potential cleavage sites for site-1 protease (S1P) that impeded the exposition of the fusion peptide for membrane fusion but reduced S-mediated infection despite normal processing levels. Conversely, N764K increased infectivity and the levels of processed S in VSVpp.
Journal reference:
- Pastorio, C., Zech, F., Noettger, S., et al. (2022). Determinants of Spike Infectivity, Processing and Neutralization in SARS-CoV-2 Omicron subvariants BA.1 and BA.2. Cell Host & Microbe. doi:10.1016/ j.chom.2022.07.006.