Encouragingly, two developed bsAbs – CoV2 biRN5 and CoV-biRN7 were found to display broad neutralization potency against a range of Omicron variants, highlighting their potential in replacing current COVID-19 intervention modalities, the latter of which are losing the arms race to SARS-CoV-2 evolution. These NTD-RBD bsAbs may form the basis for a new generation of anti-COVID-19 vaccines and antibody-based therapeutics against the disease.
 Study: Bispecific antibodies with broad neutralization potency against SARS-CoV-2 variants of concern. Image Credit: Alpha Tauri 3D Graphics / Shutterstock
Study: Bispecific antibodies with broad neutralization potency against SARS-CoV-2 variants of concern. Image Credit: Alpha Tauri 3D Graphics / Shutterstock

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Why do we need to reinvent the (COVID-19 therapeutic) wheel?
The coronavirus disease of 2019 (COVID-19) pandemic remains the worst pathogenic outbreak in recorded human history, claiming more than 7 million lives and infecting almost 800 million individuals since its discovery in China in late 2019. While government-enforced social distancing measures helped prevent the spread of the disease during the early days of the pandemic and especially during recurrent prevalence surges, most researchers agree that the single most crucial factor in eventually controlling the disease and preventing further loss of life, livelihood, and infrastructure was the rapid development and government-aided (in some cases mandated) dissemination of anti-COVID-19 vaccines.
Unfortunately, vaccines are preventive interventions against subsequent infections, not cures for patients already infected by the pathogen. In the latter cohort and for vulnerable immunocompromised individuals, a pressing need for additional clinical interventions to treat ongoing infections is required. Conventionally, monoclonal antibodies (mAbs) were developed as a safe and effective anti-COVID-19 therapy. mAbs are proteins produced by a single immune cell cohort that binds to a specific receptor on the target (herein, the SARS-CoV-2 spike protein), thereby inactivating the pathogen and allowing the body’s innate immune system to degrade or excrete the virus.
Anti-COVID-19 mAbs are derived from COVID-19 convalescent donors and, given their specificity, represent a significant improvement in efficacy over traditional broad-spectrum antivirals, with substantially fewer side effects. Unfortunately, SARS-CoV-2 is a rapidly evolving virus – the emergence of novel Omicron variants of concern (VOCs) with modified binding domains resulted in drastic mAb efficacy reductions, thereby resulting in a pressing need for the development of novel therapeutic interventions against the disease.
“We recently identified NTD-specific nAbs directed to epitopes outside the site i antigenic supersite that exhibit potent neutralization and broad binding specificity against early VOCs. Given the low immune pressure on epitopes outside the antigenic supersite on the NTD, we hypothesized that a non-NTD supersite nAb could prove effective against the latest Omicron VOCs when integrated into an NTD-RBD bsAb.”
About the study
In the present study, researchers used nAbs (especially those targeting the C1533 non-NTD supersite) derived from convalescent COVID-19 donors to develop a family of novel bispecific antibodies (bsAbs) which they then tested in vitro and in murine models against a host of novel Omicron VOCs, notably XBB.1.5, BA.2.86, and EG.5.1. They began by isolating heavy and light chain immunoglobulin G (IgGs) for each antibody and cloning them into an AbVec2.1 expression plasmid. Subsequently, these IgGs were subcloned and co-transfected into Expi293F cells for Fab construct expression. Affinity and size-exclusion chromatography were finally used to isolate and purify IgGs and Fabs of interest.
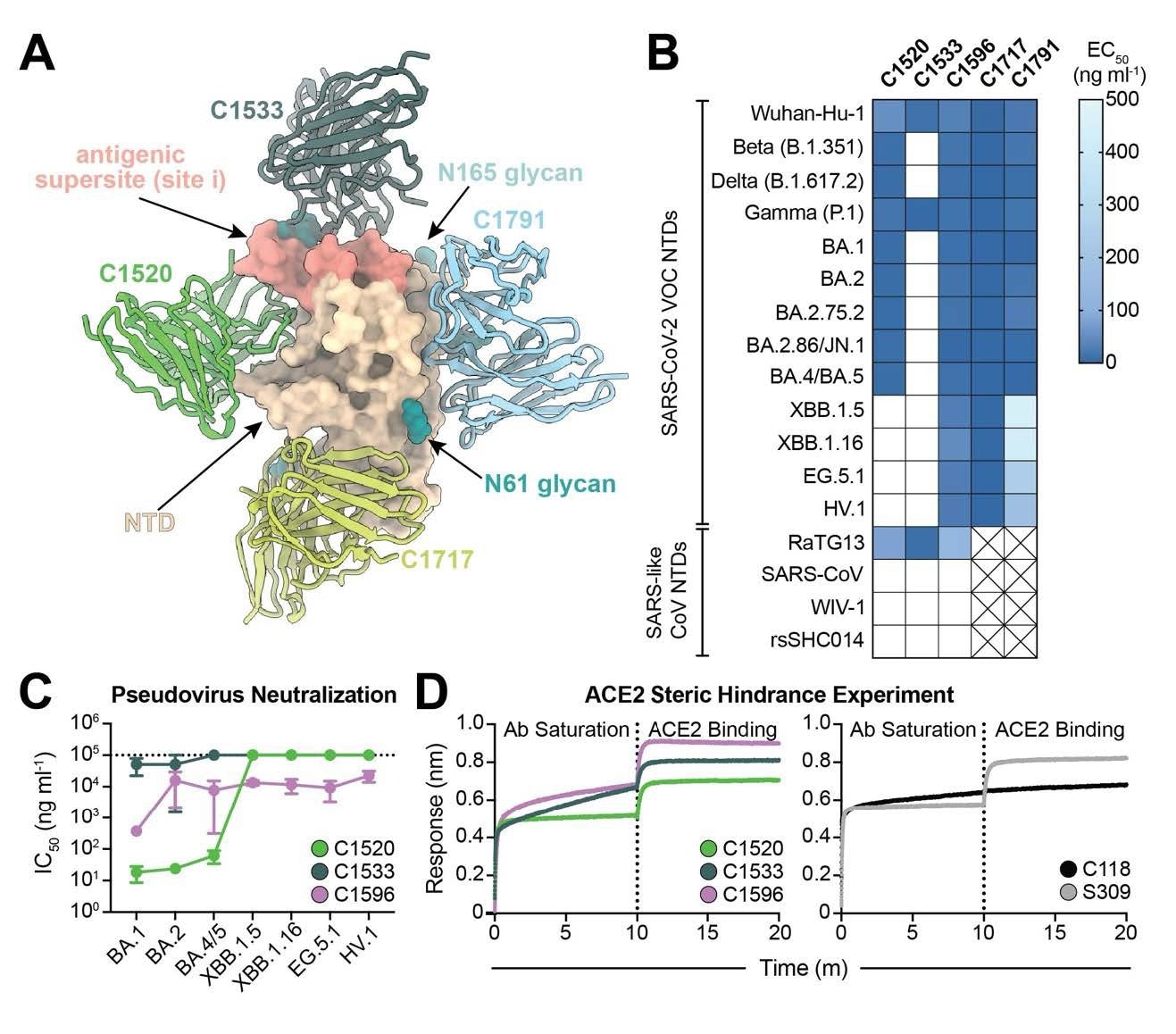
C1596, an NTD-specific mAb, exhibits broad binding and neutralizing potential across SARS-CoV-2 Omicron VOCs. (A). Superimposition of C1520 (PDB: 7UAQ, green), C1717 (PDB: 7UAR, lime-yellow), C1791 (blue), and C1533 (dark slate gray) VH and VL domains onto the SARS-CoV-2 Wuhan-Hu-1 NTD (wheat) for a composite figure. (B) Heatmap of ELISA EC50 values of NTD-specific monoclonal IgG antibodies binding to recombinant NTD protein of SARS-CoV-2 VOCs (top) and SARS-like CoVs (bottom). Boxes shaded white represent EC50 values above 500 ng ml-1; boxes with an X indicate proteins that were not tested. Data points are represented by the geometric mean of at least three independent experiments. (C) Graph with IC50 values of NTD-specific IgG antibodies against the indicated SARS-CoV-2 VOCs. Reported data are represented by the mean of at least three replicate experiments and the standard error of the mean. (D) BLI association curves for soluble human ACE2 binding to SARS-CoV-2 Wuhan-Hu-1 S 6P trimer after saturation with (left) NTD IgGs and (right) RBD IgGs. S309 IgG, an RBDspecific antibody (43) that does not compete with ACE2 for binding (43), is used as a control for successful ACE2 binding following antibody saturation of the trimer; C118 IgG, an RBD-specific antibody (23) that competes with ACE2 for RBD binding (42), is used as a control for competition with ACE2.
Researchers then developed nine bsAbs (CoV2-biRN1 to CoV2-biRN9) using a host of bsAbs-specific modalities, including the CrossMab, Xmab, dual variable domain immunoglobulin (DVD-Ig) bispecific, and tandem single-chain fragment variable (scFv) bispecific formats. Each developed bsAbs was then cloned into an AbVec2.1 expression vector and transfected into Expi293F cells for protein expression (four days at 37˚C). Enzyme-linked immunosorbent assays (ELISAs) were used to preliminarily evaluate the binding efficacy between these Abs and viral antigens. Binding kinetic data between Abs and viral antigens was measured using biolayer interferometry (BLI).
For in vitro evaluations, plasmids obtained above were transfected into HHEK293T cells for 72 hours, following which generated pseudovirions were harvested (centrifugation), filtered, and incubated for ~20 hours (37˚C and 5% CO2). For in vivo murine neutralization evaluations, K18-hACE2 transgenic mice were used (two cohorts – treatment and prophylaxis). Mice were inoculated with 2 x 104 PFU of SARS-CoV2 XBB.1.5 via intranasal administration. The treatment arm was inoculated with bsAbs 4 hours following XBB.1.5 administration while the prophylaxis arm was treated 24 hours before the viral inoculation. Three days following the experiment initiation, mice were sacrificed, and their left lung was excised for biochemical and microscopic investigations.
Study findings and conclusions
The present study reveals that the C1596 NTD-specific antibody exhibits remarkable cross-reactivity to all tested SARS-CoV-2 VOCs, suggesting that it recognizes a quaternary epitope comprising the NTD, RBD, and SD1 domains within a single spike protomer. It was found to display a substantially slower dissociation rate (kd) compared to monomeric NTD, suggesting the mechanisms unpinning its resilience to viral spike mutations. When incorporated in the bispecific format (bsAbs), six of the seven developed bsAbs using C1596 displayed significantly higher VOC neutralization efficacies than their parent mAbs in both in vitro and in vivo evaluations.
“Notably, among these constructs, CoV2-biRN5 and CoV2-biRN7 demonstrated sustained neutralization efficacy across all VOCs tested. These constructs were designed with C1596 positioned N-terminal to C952, differing from CoV2-biRN4 and CoV2-biRN6 (C952 at the N-terminus), which exhibited lower potencies. Collectively, these findings underscore the strategic importance of antibody orientation and the sequential order of binding events in designing effective bispecific therapeutics against SARSCoV-2.”
In summary, the present work underscores the benefits of bsAbs in COVID-19 infection interventions and highlights CoV-biRN5 and CoV-biRN7 as potential blueprints for developing a new generation of versatile antibodies. The authors recommend preclinical testing on these and similarly developed antibodies, paving the way for their eventual clinical translation.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
Rubio, A. A., Baharani, V. A., Dadonaite, B., Parada, M., Abernathy, M. E., Wang, Z., Lee, Y. E., Eso, M., Phung, J., Ramos, I., Chen, T., El Nesr, G., Bloom, J. D., Bieniasz, P. D., Nussenzweig, M. C., & Barnes, C. O. (2024). Bispecific antibodies with broad neutralization potency against SARS-CoV-2 variants of concern. Cold Spring Harbor Laboratory, DOI – 10.1101/2024.05.05.592584, https://www.biorxiv.org/content/10.1101/2024.05.05.592584v1