Plasmid DNA isolation is an important technique in molecular biology, however, achieving effective high-throughput automation of this method has proven to be challenging.
Analytik Jena and SynbiCITE have worked together to develop a method utilizing the CyBio® FeliX pipetting platform to isolate high-quality plasmid DNA from 96 samples in parallel, generating concentrations suitable for downstream applications such as sequencing.
The isolation of plasmid DNA from bacteria is a fundamental molecular biology technique, used in the production of template DNA for specific downstream reactions. Plasmid isolation methods are relatively straightforward, but high-throughput plasmid DNA extraction has proven difficult, with common issues such as genomic DNA (gDNA) contamination and low yield.
A multiwell-based plasmid extraction method that offers high throughput and automation, while providing a gDNA-free yield appropriate for downstream applications, will provide considerable benefits to high-throughput laboratories, where bottle-necks often occur with plasmid extraction.
96-well, silica filter-plate plasmid extraction kits can be acquired from commercial suppliers. These methods harvest bacteria cells before completing alkaline lysis. Cellular debris is then removed before the extracted DNA is collected by binding it to a silica membrane.
Membranes are then washed, before the elution of purified DNA. The method described below was developed at SynbiCITE’s London DNA Foundry. It uses the CyBio® FeliX pipetting platform to automate a protocol for 96-well, silica filter-plate-based plasmid DNA extraction.
The CyBio® FeliX pipetting platform (Figure 1) facilitates the processing of 96 samples simultaneously, in an approximately 1.5-hour period. It also offers the flexibility to process multiple plates in parallel.
.jpg)
Figure 1. Integrated Cybio® FeliX pipetting platform in SynbiCITE’s London DNA Foundry laboratory at Imperial College London.
The platform is compact, reducing the laboratory bench space required for an automated plasmid isolation system and, when used in conjunction with a benchtop robotic arm and centrifuge, it can offer a fully automated approach to plasmid extraction.
An average yield of 65 ng/µl plasmid DNA is isolated from bacterial cultures when utilizing this method, with an elution volume of 50 µl and low variability between samples (± 7.7 ng/µl SD). Plasmid DNA samples are also of high quality and ideal for downstream applications such as transformation or sequencing.
Materials and Methods
Table 1. Reagents required for method.
| Reagent |
Manufacturer |
Part Number |
| PureLinkTM Pro Quick96 Plasmid Purification Kit |
ThermoFisher Scientific |
K211004A |
| Quant-iT™ Picogreen® dsDNA Assay Kit |
ThermoFisher Scientific |
P7589 |
| Terrific Broth |
Merk |
1016290500 |
Table 2. Instrumentation used in method.
| Instrument |
Manufacturer |
| CyBio® FeliX |
Analytik Jena |
| Pipetting Head R 96/250 µl |
Analytik Jena |
| CyBio® RoboTipTray 96-250 µl DW |
Analytik Jena |
| 5810 R Centrifuge |
Eppendorf |
Table 3. Consumables required for method.
| Item |
Manufacturer |
Part Number |
| 96 well, MASTERBLOCK®, 2 ml |
Greiner |
780271 |
| 96 well plates, deep well, 1 ml |
ThermoFisher Scientific |
11381555 |
Sample Preparation
- Grow bacterial clones that contain plasmid DNA of interest
- Inoculate single clones into a 96-well, 2 ml, square-well plate that contains 1.2 ml/well Terrific Broth (TB) and appropriate antibiotic(s)
- Grow the cells overnight, shaking at 37 °C
Note that high copy number plasmids should be used to ensure increased yield.
Method
- Prepare buffers as outlined in the manufacturer’s instructions, dispensing into 96-well, 1 ml plates according to the volumes displayed in Table 4. Note that volumes provided are appropriate for 1 x 96-well plasmid extraction.
- Centrifuge the plate of overnight bacterial cultures at 2250 x g, for 10 minutes.
- Remove the supernatant, ensuring that cell pellets are kept intact.
- Add 5x RoboTipTrays, 5x buffer plates, sample plate of cell pellets, and the ‘Clarification’ plate (on top of a 96-well, 1 ml plate) to the CyBio® FeliX’s deck. This configuration is displayed in Figure 2.
Table 4. Volumes of buffer to add to 96-well, 1 ml buffer plates for 1 x 96-well plasmid extraction.
| Buffer |
Volume (µl/well) |
| Resuspension |
350 |
| Lysis |
350 |
| Neutralization |
450 |
| Wash |
1000 |
| Elution |
150 |
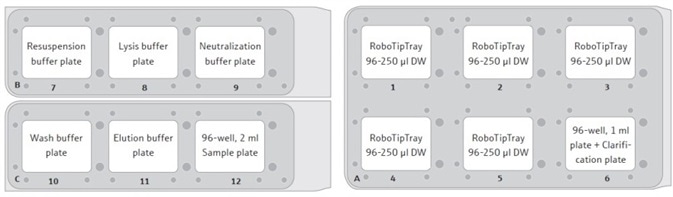
Figure 2. Suggested set-up positions for the CyBio® FeliX deck. Each RoboTipTray is used for a specific reagent(s), according to its position: (1) Resuspension buffer and sample handing (pre-clarification); (2) Lysis buffer; (3) Neutralization buffer; (4) Wash buffer and sample (post-clarifica- tion); (5) Elution buffer. The sample plate will contain cell pellets, post-harvesting by centrifugation.
- Use the CyBio® FeliX to add 250 µl/well resuspension buffer to the sample plate, then mix this thoroughly for 3 minutes using the same set of tips to resuspend the cell pellets. See Table 5 for pipetting speeds.
- Add 250 µl/well of lysis buffer to the sample plate.
- Use sample tips to mix for 4 minutes to lyse the cells.
- Add 350 µl/well of neutralization buffer to the sample plate to prevent DNA acidification.
- Mix again for 3 minutes using the sample tips.
- Use the sample tips to transfer the full volume of the lysed sample (850 µl/well) to the Clarification plate provided.
- Centrifuge the Clarification plate on top of a 1 ml, deep-well collection plate at 2250 x g for 2 minutes. The sample will pass through the clarification filter, transferring DNA to the collection plate, and removing cell debris.
- Discard the Clarification plate and return the 1 ml, deep-well collection plate to the top deck of the FeliX - position 12 – this replaces the 96-well, 2 ml sample plate.
- Add the ‘Filter’ plate provided on top of a clean 1 ml, deep-well collection plate, placing this at the bottom deck of the FeliX at position 6.
- Use clean sample tips to transfer the full volume of the clarified samples (850 µl/well) to the Filter plate.
- Centrifuge the Filter plate, on top of a 1 ml, deep-well collection plate at 2250 x g for 2 minutes. This will result in the DNA being bound to the silica membrane present in the Filter plate.
- Dispose of flow-through solution from the 1 ml, deep-well collection plate.
- Replace the Filter plate on top of the 1 ml, deep-well collection plate on the FeliX’s bottom deck at position 6.
- Use the sample tips to add 900 µl/well of wash buffer to the Filter plate. Note that the wash buffer plate must be freshly prepared fresh every time.
- Centrifuge the Filter plate, on top of the 1 ml, deep-well collection plate at 2250 x g for 2 minutes. DNA will stay bound to the silica membrane within the Filter plate.
- Dispose of the flow-through solution from the 1 ml, deep-well collection plate.
- Centrifuge the Filter plate, on top of the empty 1 ml, deep-well collection plate at 2250 x g for 10 minutes to eliminate any residual wash buffer. DNA will continue to remain bound to the silica membrane in the Filter plate.
- Place the Filter plate on top of the Elution plate provided, situating this on the FeliX’s bottom deck at position 6.
- Add 50 µl/well of elution buffer to the Filter plate.
- Incubate the plate at room temperature for 4 minutes.
- Centrifuge the Filter plate, on top of the Elution plate at 2250 x g for 2 minutes. This will elute the DNA from the silica membrane.
- Seal the Elution plate containing the purified plasmid DNA samples with a foil seal. This can be stored at 4 °C (short term) or -20 °C (long term).
Table 5. Pipetting speed for different solutions using the CyBio® FeliX pipetting platform.
| Solution |
Pipetting speed |
| Resuspension buffer |
Default |
| Lysis buffer |
15 µl/s |
| Neutralization buffer |
20 µl/s |
| Lysed sample (pre-clarification) |
15 µl/s |
| Clarified sample |
Default |
| Wash buffer II |
Default |
| Elution buffer |
Default |
Results and discussion
If a good yield from plasmid DNA isolation is to be achieved, it is essential that bacteria cells are optimally grown before the method is run. This study employed a Design of Experiments (DOE) approach to effectively optimize overnight growth of DH5α Escherichia coli (E. coli) cells at 37 °C. The extent of cell growth was established via measurement of the Absorbance of 100 µl/well culture at 600 nm (Abs600).
A custom-designed model was generated using the JMP® software, revealing that optimal cell growth conditions necessitated the use of a 2 ml, square-well, 96-well plate, which contained 60% volume of media per well (1.2 ml/well) (Figure 3 and Table 6).
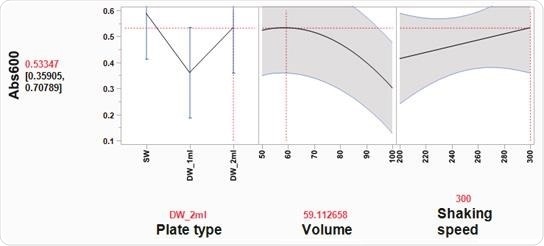
Figure 3. Optimal cell growth model visualized using the Prediction Profiler tool in JMP®. Using JMP® Design of Experiment (DOE) software, optimal growth conditions of E. coli cells in 96-well plates were determined, based on three variables: plate type, volume, and shaking speed. Three plate types were tested: SW = standard well (maximum volume 200 µl), DW_1 ml = deep, round well (maximum volume 1 ml), DW_2 ml = deep, square well (maximum volume 2 ml). Volume indicates media volume expressed as % of the maximum volume of well. DH5α E. coli cells were grown overnight at 37 °C in a shaking incubator (Kuhner ISF1-XC Climo-shaker). The Absorbance at 600 nm (Abs600) of 100 µl/well sample was measured using a BioTek SynergyTM Mx Microplate Reader. Based on the JMP® DOE model, the statistically significant effects were plate type and volume of media. In a 2 ml deep-well plate, the Prediction Profiler tool in JMP® recommends using a 60% volume of media per well (1.2 ml/well). Using these conditions the predicted Abs600 is 0.533. These conditions were tested and the actual Abs600 measured was 0.572 (± 0.12 SD)
High Abs600 values were also found where cells had been grown in a standard depth, 96-well plate. However, the reduced well volume capacity in these plates meant that the total number of cells was not adequate for successful plasmid DNA extraction. Additionally, shaking speed did not appear to have a notable impact on cell growth.
It was also determined that cell growth in Terrific Broth (TB) increased the yield of isolated plasmid DNA in comparison with Lysogeny Broth (LB) (Figure 4). The successful growth of bacteria cells under suitably optimized conditions is imperative if a good yield of plasmid DNA is to be obtained.
Under optimized conditions, the yield obtained was more than 50 ng/µl (Figure 4) in a 50 µl elution volume, which is appropriate for downstream applications such as sequencing.1
Table 6. Design of experiment (DOE) model for optimizing overnight E. coli cell growth conditions, in a 96-well plate. Three factors including plate type, the volume of media per well, and incubator shaking speed were evaluated using a custom-designed model generated with JMP® software. There were a total of 20 different runs in random orders from 5 whole plots. Each whole plot represents the same condition for shaking speed and was performed on separate days. Three plate types were tested: SW = standard well (maximum volume 200 μl), DW_1 ml = deep, round well (maximum volume 1 ml), DW_2 ml = deep, square well (maximum volume 2 ml). Volume indicates the media volume expressed as a % of the maximum volume of each well. All cells were grown overnight at 37 °C in a shaking incubator (Kuhner ISF1-XC Climo-shaker) and the response measured for each run was the Absorbance at 600 nm (Abs600) of 100 μl/well sample.
| Solution |
Pipetting speed |
Solution |
Solution |
Solution |
Solution |
| 1 |
1 |
SW |
100 |
250 |
0.242 |
| 2 |
1 |
DW_2 ml |
75 |
250 |
0.548 |
| 3 |
1 |
DW_1 ml |
100 |
250 |
0.218 |
| 4 |
1 |
DW_1 ml |
50 |
250 |
0.465 |
| 5 |
2 |
DW_2 ml |
100 |
300 |
0.195 |
| 6 |
2 |
DW_2 ml |
50 |
300 |
0.594 |
| 7 |
2 |
SW |
75 |
300 |
0.552 |
| 8 |
2 |
DW_1 ml |
75 |
300 |
0.172 |
| 9 |
3 |
SW |
50 |
250 |
0.484 |
| 10 |
3 |
DW_1 ml |
50 |
250 |
0.24 |
| 11 |
3 |
DW_2 ml |
75 |
250 |
0.49 |
| 12 |
3 |
DW_1 ml |
100 |
250 |
0.117 |
| 13 |
4 |
SW |
75 |
200 |
0.465 |
| 14 |
4 |
DW_2 ml |
50 |
200 |
0.265 |
| 15 |
4 |
DW_1 ml |
75 |
200 |
0.182 |
| 16 |
4 |
DW_2 ml |
100 |
200 |
0.127 |
| 17 |
5 |
DW_1 ml |
75 |
250 |
0.176 |
| 18 |
5 |
SW |
50 |
250 |
0.507 |
| 19 |
5 |
SW |
100 |
250 |
0.37 |
| 20 |
5 |
DW_2 ml |
75 |
250 |
0.46 |
-1.jpg)
Figure 4. Optimal growth media for plasmid DNA isolation from E. coli cells. E. coli DH5α cells were grown overnight, shaking at 37 °C, in a 2 ml, square-well, 96-well plate in either Lysogeny broth (LB) or Terrific Broth (TB), 1200 µl/ well. Plasmid DNA (p15a ORI) was extracted from the cells and the concentration of the isolated DNA was determined using a NanoDrop (Thermo). The average data from three replicates are plotted with error bars representing the standard deviation (SD). The isolated DNA yield from cells grown overnight in TB media was greater than when cells were grown in LB. Furthermore, the average yield from cells grown in TB was greater than 50 ng/µl in a 50 µl elution volume, which is sufficient for sequencing. The average 260/280 ratio was 1.9 (±0.07 SD) and 1.97 (±0.07 SD) for cells grown in LB and TB respectively, indicating good DNA purity.
The plasmid isolation method outlined above was found to provide a reproducible yield from samples that were processed simultaneously in a 96-well plate (Figure 5).
h.jpg)
Figure 5. Plasmid DNA extracted from multiple bacterial E. coli samples simultaneously, using an automated method on the CyBio® FeliX. Using the automated method described here, plasmid DNA (p15a ORI) was isolated from E. coli DH5α cells, grown under optimized conditions. The isolated plasmid DNA yield was reproducible across the samples with an average yield of 65.3 ng/μl (± 7.7 ng/μl SD). The average 260/280 ratio was 2.0 (± 0.05 SD), indicative of good DNA quality.
High-quality DNA that provides an average yield of 65.3 ng/µl (+/- 7.7 SD) in a 50 µl volume can be obtained from a low copy number plasmid (p15a ORI). This is adequate for downstream applications, including transformation into bacteria cells, or sequencing.
As anticipated, utilizing a higher copy number plasmid was found to generate a higher yield of isolated plasmid DNA (Figure 6), meaning that the use of high copy number plasmids presents an opportunity for higher yield of plasmid DNA where necessary.
.jpg)
Figure 6. The effect of plasmid copy number on isolated DNA yield. DH5α E. coli cells were transformed with cloning vectors with different origins of replication (ORI) to investigate the effect of copy number on the yield of isolated plasmid DNA. p15α plasmids have a low copy number while ColE1 plasmids have a higher copy number. The average data from three replicates are plotted with error bars representing the standard deviation (SD). The use of the higher copy number plasmid, ColE1, increases the isolated plasmid DNA yield by more than 2-fold, as compared to the p15a plasmid. The average 260/280 ratios were 1.91 (± 0.08 SD) and 1.89 (± 0.03 SD) for p15a and ColE1 plasmids respectively, indicating good DNA quality.
Using the CyBio® FeliX pipetting system, this method can be utilized to process a multiwell plate of 96 samples in parallel. If this method is used in combination with a robotic arm and an integrated centrifuge with appropriate filter plate depth, it is possible to fully automate and adapt the method for processing multiple 96-well sample plates simultaneously.
Conclusion
Demand for higher throughput in molecular biology laboratories continues to increase, meaning that essential techniques such as plasmid DNA isolation must be implemented on a high-throughput, automated scale.
This article outlined a method designed to enable high-throughput isolation of plasmid DNA from bacterial cell cultures, which has the potential to be fully automated. The method was shown to isolate quantities of high-quality, plasmid DNA from bacterial cultures, at yields that are more than adequate for downstream applications.
DNA yield is very much dependent on how successful the overnight growth of bacterial cultures is, so optimized growth conditions are vital if this method is to be successful.
The compact CyBio® FeliX pipetting platform has been specifically developed for high-throughput plasmid DNA isolation, offering a viable option for molecular biology laboratories that need to meet demand while avoiding this common bottleneck in laboratory workflow.
References and Further Reading
- https://eurofinsgenomics.eu/
Acknowledgments
In cooperation with SynbiCITE
About Analytik Jena US
Analytik Jena is a provider of instruments and products in the areas of analytical measuring technology and life science. Its portfolio includes the most modern analytical technology and complete systems for bioanalytical applications in the life science area.
Comprehensive laboratory software management and information systems (LIMS), service offerings, as well as device-specific consumables and disposables, such as reagents or plastic articles, complete the Group’s extensive range of products.
About life science
The Life Science product area demonstrates the biotechnological competence of Analytik Jena AG. We provide a wide product spectrum for automated total, as well as individual solutions for molecular diagnostics. Our products are focused to offer you a quality and the reproducibility of your laboratory results.
This will surely ease your daily work and speed up your work processes in a certain way. All together we support you through the complete process of the lab work. Besides we offer customized solutions and are able to adapt our products to your needs. Automated high-throughput screening systems for the pharmaceutical sector are also part of this segment’s extensive portfolio.
About analytical instrumentation
Analytik Jena has a long tradition in developing high-performance precision analytical systems which dates back to the inventions made by Ernst Abbe and Carl Zeiss. We have grown to become one of the most innovative manufacturers of analytical measuring technology worldwide.
Our business unit Analytical Instrumentation offers excellent competencies in the fields of optical spectroscopy, sum parameters and elemental analysis. Being proud of our core competency we grant all our customers a long-term warranty of 10 years for our high-performance optics.
About lab automation
With more than 25 years of market experience, Analytik Jena with its CyBio® Product Line is a leading provider for high quality liquid handling and automation technologies. In the pharmaceutical and life science industries, our products enjoy the highest reputation for precision, reliability, robustness and simplicity.
Moreover, the Automation Team designs, produces and installs fully automated systems tailored to our clients' application, throughput and capacity requirements. From stand-alone CyBio® Well up to fully customized robotic systems we handle your compounds, biomolecules and cells with great care.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.