Sample volumes that are gradually increasing and innovative options for molecular biology experiments are altering the demands on quantitative real-time PCR. Migrating to the multi-well format with 384 samples creates new application opportunities for gene expression, mutation analysis, and diagnostics.

A steady homogeneous readout of minute sample volumes is needed for real-time PCR in 384-well high-throughput format. In addition to factors such as speed and experimental accuracy in the high-throughput format, which in turn, will play a progressively important role.
The qTOWER³ 84 delivers a performance that is precise, and is shown in 384-well high-throughput format. This is done through an aluminum block that guarantees temperature consistency. Furthermore, the patented high performance optics allow for a readout time of merely six seconds for a complete 384-well plate, regardless of the amount of fluorescent dyes that are used.
Something to consider is that the preparation requirements to enable high-throughput qPCR performance is steadily increasing. Moreover, the system setup for the reaction batch for qPCR needs specific expertise and skills. In addition to quadrupling the amount of samples of the 96-well standard format, the decreased sample volume also plays a significant role.
Extreme precision is extremely important during the pipetting of these micro-volume ranges. If this is not completed, even the tiniest disparities in raw material can result in huge variances in the amplification.
Methods and Materials Used
A sequence-specific primer pair and the double-concentrated innuMIX qPCR MasterMix SyGreen (Analytik Jena) DNA is extracted from Escherichia coli. In the 384-well format, an E.coli-specific target sequence of 120 bp is amplified in real-time.
| A) Manual |
B) Automated |
| Reaction volume |
Ct |
SD(Ct) |
Reaction volume |
Ct |
SD(Ct) |
| 2 µL |
14.926 |
0.05 |
2 µl |
14.896 |
0.03 |
| 5 µL |
14.464 |
0.04 |
5 µl |
14.430 |
0.02 |
| 10 µL |
14.128 |
0.05 |
10 µl |
14.168 |
0.02 |
| 20 µL |
14.12 |
0.08 |
20 µl |
14.096 |
0.04 |
Figure 1. Comparison of the automatically generated Ct values of the qPCRsoft in the qTOWER3 84.
Five technical replicas, namely 5 µl to 20 µl, of the master mix were used in a variation of the reaction volume. As a result, the specificity of the product amplification was confirmed in a melting curve. FrameStar® qPCR plate (4titude) carried out the amplification, with a preliminary denaturing of 120 s followed by 35 cycles, with the denaturation for 15 s at 95 degrees Celsius, annealing for 15 s at 58 degrees Celsius, followed by elongation for 30 s at 72 Celsius.
In each cycle, the fluorescence signal was recorded at 72 degrees Celsius. A comparison was made between the completion of a manual reaction setup with the use of a manual pipette, and completion of an automated reaction batch that was done using the pipetting robot GeneTheatre (Analytik Jena).
Results and Discussion
The volume variation results demonstrate that there is a possibility to decrease the reaction volume up to 2 µl for this assay while not compromising reproducibility and precision. At a sample volume of just this small size, there is a standard deviation of the Ct values of only ± 0.05 over five repetitions.
Results show the amplification line plot from the automatically generated reaction setup using the pipetting robot indicating a homogeneous distribution of the volume-varied real-time PCR curves.
The software calculated the standard deviation of the Ct values which are yet again, lower in comparison to the results of the manual pipette Ct values in the experiment.
Consequently, it is explained that validated and reproducible results are attained using a pipetting robot, especially in the 384-well high-throughput format, through standardization of the sample setup and the pipetting routine. This is seen even in the lowest volume range.
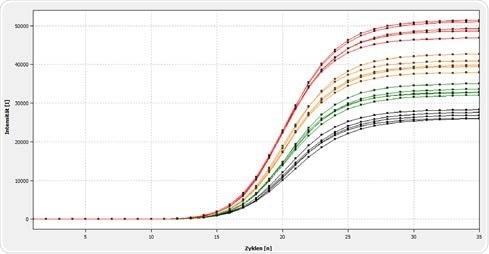
Figure 2a. Representation of the amplification of a 120 bp E.coli-specific target sequence in the qTOWER3 84. In the image above, a manual reaction setup has been used, using the manual pipette for the carrying out the volume variation.
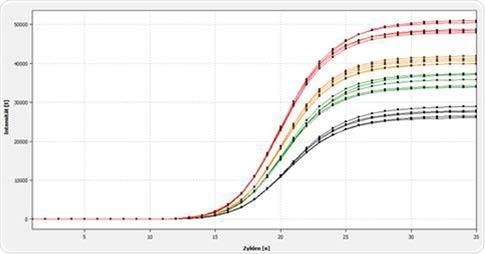
Figure 2b. Representation of the amplification of a 120 bp E.coli-specific target sequence in the qTOWER3 84. At this point, the qPCR was carried out with an automated reaction batch, produced using the GeneTheatre pipetting robot.
Conclusion
Using a pipetting robot for standardizing and improving the precision and reproducibility of realtime PCR reactions is highly recommended. It makes it substantially easier to complete all pipetting tasks that are pending in the laboratory, and enables complete automation of PCR and real-time PCR reaction batches.
The standardization of processes significantly reduces errors regarding reactions involving 10 or 5 µl batch volumes – regardless of the skill or experience of the user.
An automated reaction setup provides a range of advantages: the utmost precision, e.g., as well as the possibility to routinely process smaller reaction volumes than those which would be possible with manual batches.
References
Labo 5/2017: Kleinstvolumina im Hochdurchsatz messen
About Analytik Jena US
 Analytik Jena is a provider of instruments and products in the areas of analytical measuring technology and life science. Its portfolio includes the most modern analytical technology and complete systems for bioanalytical applications in the life science area.
Analytik Jena is a provider of instruments and products in the areas of analytical measuring technology and life science. Its portfolio includes the most modern analytical technology and complete systems for bioanalytical applications in the life science area.
Comprehensive laboratory software management and information systems (LIMS), service offerings, as well as device-specific consumables and disposables, such as reagents or plastic articles, complete the Group’s extensive range of products.
About Life Science
The Life Science product area demonstrates the biotechnological competence of Analytik Jena AG. We provide a wide product spectrum for automated total, as well as individual solutions for molecular diagnostics. Our products are focused to offer you a quality and the reproducibility of your laboratory results.
This will surely ease your daily work and speed up your work processes in a certain way.
All together we support you through the complete process of the lab work. Besides we offer customized solutions and are able to adapt our products to your needs. Automated high-throughput screening systems for the pharmaceutical sector are also part of this segment’s extensive portfolio.
About Analytical Instrumentation
Analytik Jena has a long tradition in developing high-performance precision analytical systems which dates back to the inventions made by Ernst Abbe and Carl Zeiss. We have grown to become one of the most innovative manufacturers of analytical measuring technology worldwide.
Our business unit Analytical Instrumentation offers excellent competencies in the fields of optical spectroscopy, sum parameters and elemental analysis. Being proud of our core competency we grant all our customers a long-term warranty of 10 years for our high-performance optics.
About Lab Automation
With more than 25 years of market experience, Analytik Jena with its CyBio® Product Line is a leading provider for high quality liquid handling and automation technologies. In the pharmaceutical and life science industries, our products enjoy the highest reputation for precision, reliability, robustness and simplicity.
Moreover, the Automation Team designs, produces and installs fully automated systems tailored to our clients' application, throughput and capacity requirements. From stand-alone CyBio® Well up to fully customized robotic systems we handle your compounds, biomolecules and cells with great care.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.