The process of bacterial transformation involves the introduction of foreign plasmid DNA into a bacterial cell. The transformation of DNA into bacteria cells, for example, Escherichia coli (E. coli), is an important molecular biology method used for the analysis of bacteria itself as well as for the storage and/or replication of plasmids. To facilitate bacterial transformation, DNA plasmids are usually designed to include a bacterial origin of replication (ORI) and an antibiotic resistance gene, to be used as a selection marker.
The use of heat shock is one easy technique for the conversion of plasmid DNA into bacteria cells. Plasmid DNA is incorporated into potential bacteria cells and incubated in the cold. The cells are subsequently heated to a high temperature for a short period of time prior to being returned to the cold. This “heat shock” allows the transfer of the plasmid DNA within the bacteria cell.
Following recovery in media, cells consisting of the plasmid DNA of interest are ready to be cultured, normally on solid agar by adding antibiotics to the agar. Antibiotic selection can be employed to choose those cells which have been successfully transformed with DNA.
The transformation of bacteria cells usually remains a manual, low-throughput activity in molecular biology laboratories. The potential to modernize bacteria transformation into a high-throughput, automated one would significantly help laboratories seeking to increase their throughput and enhance the reproducibility and robustness of their experiments. The CyBio® FeliX provides a compact pipetting platform for carrying out the liquid handling steps needed for a high-throughput transformation technique.
It is possible to completely automate the transformation method by using it on an automation platform together with a thermal cycler, a heating/cooling block, a shaking microplate-compatible incubator, and a robotic arm (Figure 1). The use of the CyBio® FeliX has allowed SynbiCITE to produce a fully automated, multi-well plate based technique for the concurrent transformation of 96 DNA plasmids into bacteria cells by heat shock, and the subsequent plating and development of these transformed cells on solid agar. Furthermore, the technique can be easily executed to process multiple plates simultaneously by using extra heating/cooling blocks.

Figure 1. Automation platform, including a Cybio® FeliX pipetting platform, in SynbiCITE’s London DNA Foundry laboratory at Imperial College London.
Challenge
The automation of the process of bacterial transformation in molecular biology laboratories.
Solution
A completely automated technique for the high-throughput transformation and growth of bacteria cells with the help of the CyBio® FeliX.
Materials and Methods
Reagents and Instrumentation
Table 1. Reagents required for the method
| Reagent |
Manufacturer |
Part Number |
| Super Optimal broth with Catabolite repression (SOC) |
Formedium |
SOC0202 |
| Lysogeny broth (LB) agar |
Merck |
1102850500 |
Table 2. Instrumentation used in the method
| Instrument |
Manufacturer |
| CyBio® FeliX |
Analytik Jena |
| Pipetting Head R 96/250 μl |
Analytik Jena |
| CyBio® RoboTipTray 96-250 μl DW |
Analytik Jena |
| TRobot Thermocycler |
Analytik Jena |
| CPAC Ultraflat heating/cooling block |
Inheco |
| CoolRack® XT PCR96 |
BioCision |
| StoreX shaking incubator |
LiCONiC Instruments |
| PreciseFlex Robot |
Precise Automation |
Table 3. Consumables used in the method
| Consumables |
Manufacturer |
Part Number |
| 96-well, F-bottom plate |
Greiner |
781201 |
| 96-well, PCR plate |
4titude |
4ti-0960 |
| 384-well, PCR plate |
4titude |
4ti-0384 |
| OmniTray |
Nunc |
242811 |
Sample Preparation
- A 384-well PCR plate consisting of plasmid DNA samples (>10 µL/well) is prepared. Care must be taken to ensure that a positive control plasmid is taken, which comprises of the right antibiotic resistance gene(s). It should be noted that DNA will be transferred from the 384-well source PCR plate to a 96-well destination plate, hence samples must be added to the 384-well plate appropriately (Figure 2).
- 20 µL/well competent E. coli cells are covered in a hard-shell 96-well, PCR plate. They are stored at −80 °C before use
- Lysogeny broth (LB) solid agar containing the suitable antibiotics for selection is prepared and placed in an OmniTray, 30 mL/plate
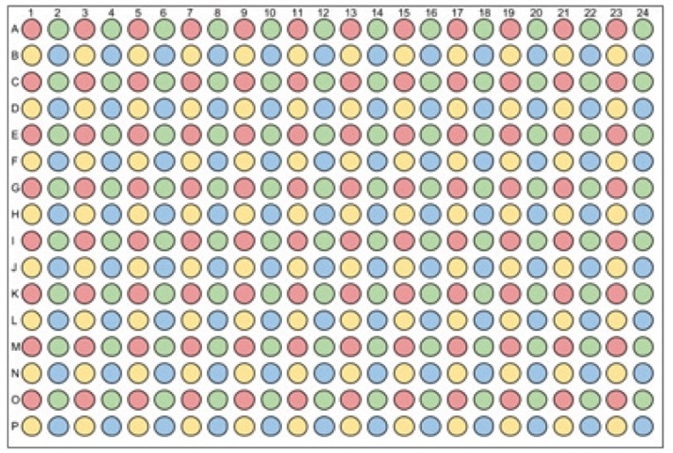
Figure 2. 384-well PCR DNA source plate. DNA will be transferred from a 384-well PCR source plate to a 96-well destination plate. One 384-well PCR DNA source plate can be used to store DNA for up to four 96-well transformations. To enable transfer to a 96-well plate, DNA should be added to the 384-well plate in quadrants: red = quadrant A1; green = quadrant A2; yellow = quadrant B1; blue = quadrant B2.
Method
- A 96-well, flat-bottom plate comprising of 120 µL/well Super Optimal broth with Catabolite repression (SOC) is heated to 37 °C
- The 2x CyBio® RoboTipTray 96–250 µL DW is added to the deck of the CyBio® FeliX (Figure 3)
- The 384-well plasmid DNA source plate is added to the deck of the CyBio® FeliX (Figure 3)
- A 96-well PCR plate having 20 µL/well competent E. coli cells is removed from −80 °C and placed immediately onto a CoolRack® XT PCR96, which has been cooled to 4 °C
- The CoolRack® (with the cell plate) is placed on to the heating/cooling block set to 4 °C and cells are thawed for 10 minutes. It is important to make sure that no moisture is present on the surface of the heating/cooling block and/or the base of the CoolRack®, before use. The presence of moisture will stop the robot from being able to pick up CoolRack® in the following steps.
- With the CyBio® FeliX, 5 µL/well plasmid DNA is aspirated from the source plate and the volume is held in the tips
- The cell plate, still in the CoolRack®, is transferred from the heating/cooling block to the deck of the CyBio® FeliX robot (Figure 3) and the 5 µL plasmid DNA is dispensed right away
- The CoolRack® (with the cell plate) is rapidly returned to the heating/cooling block, configured at 4 °C
- The cells, with DNA, are incubated for 30 minutes at 4 °C
- Using a thermal cycler, the cells are subjected to a heat shock for 45 seconds at 43 °C. It should be noted that the time and temperature are optimized independently for each automation platform.
- The cells are immediately replaced on the CoolRack® and incubated for 2 minutes at 4 °C
- The cells are transferred to the deck of the CyBio® FeliX (Figure 3)
- The warmed 96-well, F-bottom plate consisting of SOC is transferred to the deck of the CyBio® FeliX (Figure 3)
- An 80-µL/well SOC is aspirated and added to the cell plate
- The cells are suspended again in the warmed SOC by mixing for one time
- The whole volume of SOC/cells is transferred to the 96-well, F-bottom SOC plate
- The 96-well, F-bottom SOC/cells plate is closed with a lid and incubated at 37 °C, for 60 minutes, and shaken
- In the course of the incubation, the OmniTray plate, having the solid LB agar, is warmed to 37 °C, by taking away the lid, to dry the agar before the plating of the cells. Care must be taken to ensure that the incubator has low humidity to enable drying, since drying time will change according to the incubator but may take the complete 60 minutes.
- The 96-well plate, consisting of the SOC recovered cells, and the LB agar plate from the incubator are returned to the deck of the CyBio® FeliX (Figure 3)
- Using a new set of tips, the SOC cell samples are mixed and 3-µL/well is aspirated
- The cell solution is dispensed onto the surface of the LB agar. It should be noted that the height is set such that the tips touch the agar surface but do not penetrate
- The samples are allowed to dry on the surface of the agar (around 2–3 minutes)
- The transfer of the samples to the surface of the LB agar is repeated for the required number of times, leaving adequate drying time between “spotting” events. The CyBio® FeliX method can be set up to allow these parameters to be specified by the user at the start of the protocol.
- The LB agar plate is closed with lids and incubated overnight in a humidified incubator at 37 °C, to facilitate colony growth. If needed, a reservoir containing H2O is incorporated to the incubator to maintain humidity and to prevent splitting of agar.
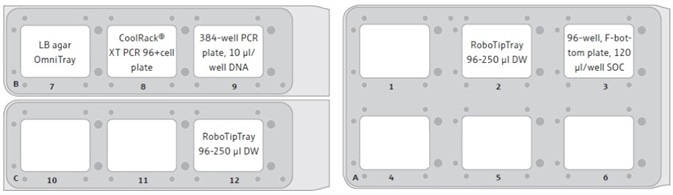
Figure 3. CyBio® FeliX deck layout for the automated transformation method. Here, an example of the FeliX deck layout is shown, as used in the London DNA Foundry. After the DNA addition step, the 384-well PCR plate containing the DNA is transferred to an accessible position on the automation platform, allowing for the safe removal of the plate from the platform and its subsequent storage for potential future use. After the transfer of the cells from the 96-well PCR plate to the 96-well, F-bottom SOC plate, the CoolRack® (with cell plate) can be removed from the FeliX deck to free up position 8. The empty spaces (blank) represent available positions, which allow for the simple expansion of the method to process multiple DNA/cell/LB agar plates at a time.
Results and Discussion
The transformation efficiency of E. coli cells was identified to be highly sensitive to the temperature of the thermal cycler, used for the heat shock. The optimal temperature will probably differ between various machines and laboratories and therefore must be determined independently by any group using this technique. A heat shock at 43 °C for 45 seconds offered the optimal transformation of DNA into E. coli cells using the TRobot Thermocycler in the London DNA Foundry. For lower concentrations of plasmid DNA, the transformation efficiency was considerably enhanced at 43 °C than when cells were heated to 42 °C for the same time period (Figure 4).
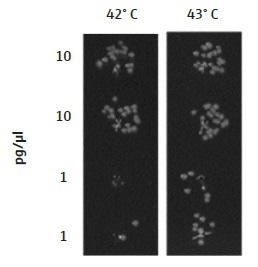
Figure 4. Temperature of the TRobot Thermocycler for the heat shock step. The optimal temperature for the heat shock of bacterial cells will be dependent on the equipment being used and should be optimized for each laboratory independently. A difference in 1° C can have a great effect on transformation efficiency, as seen here where cells were heated to either 42 °C or 43 °C. In the London DNA Foundry, 43 °C was the optimal temperature for a 45-second heat shock, showing greater transformation efficiency of lower concentrations of DNA (1 pg/µL), as compared to 42 °C. Data are shown for DH5α E. coli cells transformed with a plasmid containing Chloramphenicol and Kanamycin resistance genes (p15a ORI).
The importance of maintaining the competent E. coli cells at 4 °C always after taking them away from the −80 °C freezer, before the heat shock step, is demonstrated. Removing the cells from 4 °C for less than 2 minutes for the DNA addition step leads to a considerable decrease in the transformation efficiency of the cells (Figure 5). To maintain the cells at 4 °C at the time of the automated addition of DNA on the CyBio® FeliX, a CoolRack® XT PCR96 (BioCision) was chosen to use. The CoolRack® is first cooled to 4 °C in the refrigerator, before use. Competent E. coli cells are transferred to the CoolRack® instantly after removal from −80 °C and the CoolRack® temperature is maintained at 4° C by placing it on a heating/cooling block, added to the automation platform, and set to 4 °C.
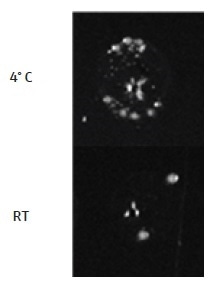
Figure 5. Temperature of cells for DNA addition step. It is very important for competent E. coli cells to be kept at 4° C at all times prior to the heat shock step. Shown here are DH5α cells which were either maintained at 4 °C or moved temporarily to the CyBio® FeliX deck at room temperature (RT; <2 minutes), for the DNA addition step. The transformation efficiency becomes very low when the DNA addition is carried out at room temperature. Therefore, during the DNA addition step on the CyBio® FeliX, E. coli cells must be kept at 4 °C. This can be achieved by placing the cell plate on a CoolRack® XT PCR96, cooled to 4 °C, and transferring the CoolRack® (containing the cell plate) to the CyBio® FeliX deck for the addition of the DNA.
The CoolRack®, with the cell plate on top, is transferred to the deck of the CyBio® FeliX to add the DNA, prior to being immediately replaced on the heating/cooling block. This maintains the cells at an adequately low temperature for excellent transformation efficiency. In order to reduce the time the CoolRack® and cells are off the heating/cooling block, the CyBio® FeliX is configured to aspirate the DNA before the robot moves the CoolRack® and cells from the heating/cooling block to the deck of the CyBio® FeliX.
The efficiency of this automated bacterial transformation technique is adequate for use with plasmids with a low copy number, for example, those with the p15a ORI (Figure 4). However, the use of a plasmid vector comprising of an ORI with a high copy number, for example, pUC19, produces a greater number of colony forming units, when compared to a plasmid with a lower copy number, for example, pBBR1 (Figure 6) or p15a (Figure 4).
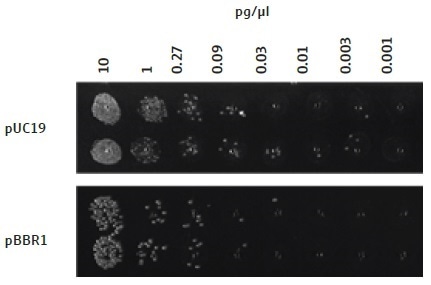
Figure 6. Transformation of E. coli cells with plasmids of different copy number. As expected, the copy number of a plasmid affects the number of colonies forming units observed after overnight growth. A higher copy number plasmid, pUC19, gives rise to a larger number of colonies as compared to a medium copy number plasmid, pBBR1, when the same concentration of DNA is added to the cells.
Thus, the plasmid copy number must be considered while deciding the concentration of DNA to convert into cells and also while determining the amount of transformed sample to plate onto solid agar for the overnight growth step. This technique can also be employed to effectively transform plasmid DNA into various strains of competent E. coli cells, which are acquiescent to heat shock transformation (Figure 7).
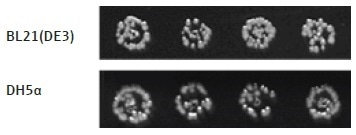
Figure 7. Different E. coli strains transformed with a positive control plasmid. The transformation protocol can be used to effectively transform different strains of E. coli cells with plasmid DNA. Here, two E. coli strains, BL21(DE3) and DH5α, have been transformed with the same plasmid DNA, containing Chloramphenicol and Kanamycin resistance genes (p15a ORI).
It is recommended to use transformation controls when running this technique. As a positive control, a plasmid DNA with the same antibiotic resistance gene as the test plasmids must be used. Replicates are recommended to supervise the variability of the transformation efficiency between wells (Figure 8). As a negative control, a number of wells of the cell plate must have no DNA added.
When these cells are cultured on solid agar containing antibiotics, there must not be any bacterial growth in these positions (Figure 8). The automated method explained in this article involves utilizing the CyBio® FeliX robot to allow the simultaneous transformation of 96 different DNA plasmids into E. coli cells by heat shock and their following plating onto agar. The outcome is an agar plate having bacterial colonies cultured in specific different regions, according to a standard 96-well reference, from which individual clones can be chosen and cultured for further analysis (Figure 8).
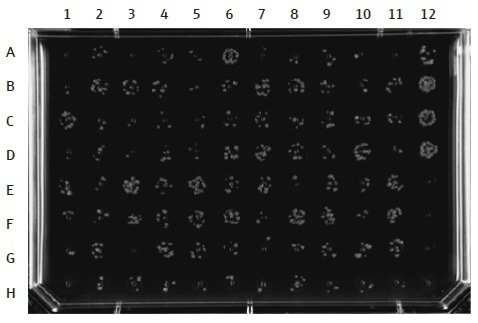
Figure 8. DH5α E. coli clones, grown in a 96-well layout on a single LB agar plate after an automated 96-well transformation with plasmid DNA. The automated transformation method can be used to transform 96 plasmid DNA samples simultaneously. Controls should be included in each transformation experiment. Here, column 12 is used for the controls: A–D = positive control plasmid (with appropriate antibiotic resistance genes); E–H = negative control (no DNA added).
The technique mentioned in this article is suitable for the complete automation of bacterial transformation and colony growth. However, the protocol can be easily tailored for use on a standalone CyBio® FeliX pipetting platform. Together with the manual completion of some steps, the use of a standalone CyBio® FeliX would still offer a high-throughput and robust technique for bacterial transformation and cell plating, in laboratories that do not possess a completely automated platform.
Conclusion
The potential to carry out high-throughput, automated bacterial transformations would be of great use to laboratories that wish to increase their throughput and enhance the reproducibility and robustness of their experiments. In this article, a fully automated technique is described for the transformation of plasmid DNA into E. coli cells by heat shock, using the CyBio® FeliX robot pipetting platform.
It has been demonstrated that this technique can be used to efficiently transform 96 DNA plasmids into various E. coli strains at the same time and is appropriate for plasmids with ORIs ranging from low to high copy number. The use of the compact CyBio® FeliX as part of an incorporated robotic platform offers an option for complete automation of the procedure. On the other hand, the method used on a standalone CyBio® FeliX pipetting platform, together with manual completion of some steps, would still offer a robust and high-throughput technique for cell plating and bacterial transformation.
About Analytik Jena US
Analytik Jena is a provider of instruments and products in the areas of analytical measuring technology and life science. Its portfolio includes the most modern analytical technology and complete systems for bioanalytical applications in the life science area.
Comprehensive laboratory software management and information systems (LIMS), service offerings, as well as device-specific consumables and disposables, such as reagents or plastic articles, complete the Group’s extensive range of products.
About Life Science
The Life Science product area demonstrates the biotechnological competence of Analytik Jena AG. We provide a wide product spectrum for automated total, as well as individual solutions for molecular diagnostics. Our products are focused to offer you a quality and the reproducibility of your laboratory results.
This will surely ease your daily work and speed up your work processes in a certain way. All together we support you through the complete process of the lab work. Besides we offer customized solutions and are able to adapt our products to your needs. Automated high-throughput screening systems for the pharmaceutical sector are also part of this segment’s extensive portfolio.
About Analytical Instrumentation
Analytik Jena has a long tradition in developing high-performance precision analytical systems which dates back to the inventions made by Ernst Abbe and Carl Zeiss. We have grown to become one of the most innovative manufacturers of analytical measuring technology worldwide.
Our business unit Analytical Instrumentation offers excellent competencies in the fields of optical spectroscopy, sum parameters and elemental analysis. Being proud of our core competency we grant all our customers a long-term warranty of 10 years for our high-performance optics.
About Lab Automation
With more than 25 years of market experience, Analytik Jena with its CyBio® Product Line is a leading provider for high quality liquid handling and automation technologies. In the pharmaceutical and life science industries, our products enjoy the highest reputation for precision, reliability, robustness and simplicity.
Moreover, the Automation Team designs, produces and installs fully automated systems tailored to our clients' application, throughput and capacity requirements. From stand-alone CyBio® Well up to fully customized robotic systems we handle your compounds, biomolecules and cells with great care.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.