In this interview, Chris Adams Ph.D., Business Development Director, Global Bioinformatics, Life Science Mass Spectrometry at Bruker Daltonics, Billerica, MA, USA talks to News-Medical Life Sciences about acquiring real-time, accurate and reproducible 4D-proteomics data with PaSER.
Could you start by giving our readers a brief introduction to the PaSER platform?
PaSER (Parallel Search Engine in Real-Time) is our real-time bioinformatics solution. PaSER performs database searching in parallel with data acquisition on a high-speed timsTOF mass spectrometer. It has been designed to offer a complete ‘run and done’ experience and to streamline workflows maximizing the effciency of turning data to usable information.
PaSER was initially launched a year and a half ago. It is essentially an independent GPU-enabled box that sits adjacent to your acquisition PC. It runs on an Nvidia graphics card with more than 4,000 programable cores, meaning it is able to search data at the speed of acquisition, including PTMs. Offline searches also benefit from the power of the GPU.
Information coming from the acquisition PC, such as retention time, ion mobility, precursor m/z and fragment ion spectra, is all sent to PaSER and processed in real-time. Database search results are delivered as soon as the acquisition is done – that is why we call it ‘run and done.’
It is important to point out that to ensure a viable real-time search at 120 Hz scan speed, we need to be achieving a per-spectra search time of less than 8 ms.
The GPU-enabled search is the brainchild of Robin Park, who developed a way to parallelize processing across a GPU. Instead of searching across tens of CPU cores, you are actually searching across thousands of GPU cores.
This expedites the search substantially, and we can do this with PTMs and in some semi-tryptic modification settings, depending on database size.
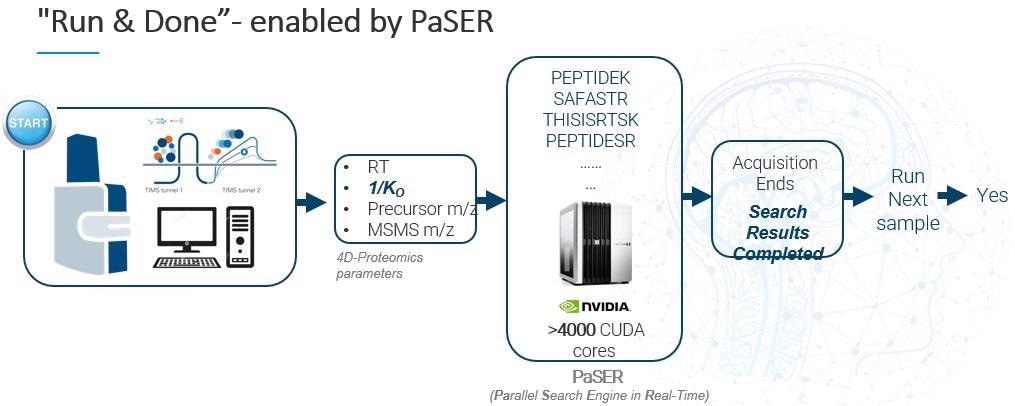
Image Credit: Bruker Daltonics
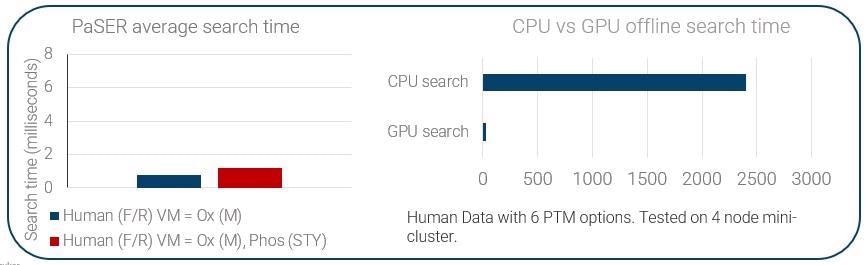
Image Credit: Bruker Daltonics

Image Credit: Bruker Daltonics
How does TMT Quant work in PaSER, and what are some of the advantages of using this method?
When Bruker first launched its timsTOF Pro, there was some skepticism around its ability to do TMT, but you can do TMT with this instrument with added benefits of sequencing speed and reduced interference.
There are some limitations around resolution at the lower m/z levels but you can do TMT 9 or 10-plex as others have shown and you can search that on the PaSER box in near real-time.
General search parameters such as enzymes and static modifications are typically set before data acquisition, meaning that real-time identification is possible. The parameters are fully customizable, and these can be set for any set of reporter ions being used.
Once the database search is done, the user will select quantitative analysis, set any options relating to the reporter ions used and desired tolerances and simply click ‘submit’ to get results.
In terms of its quantification accuracy and efficiency, this approach is as good, if not better, than other packages accepted as the gold standard.
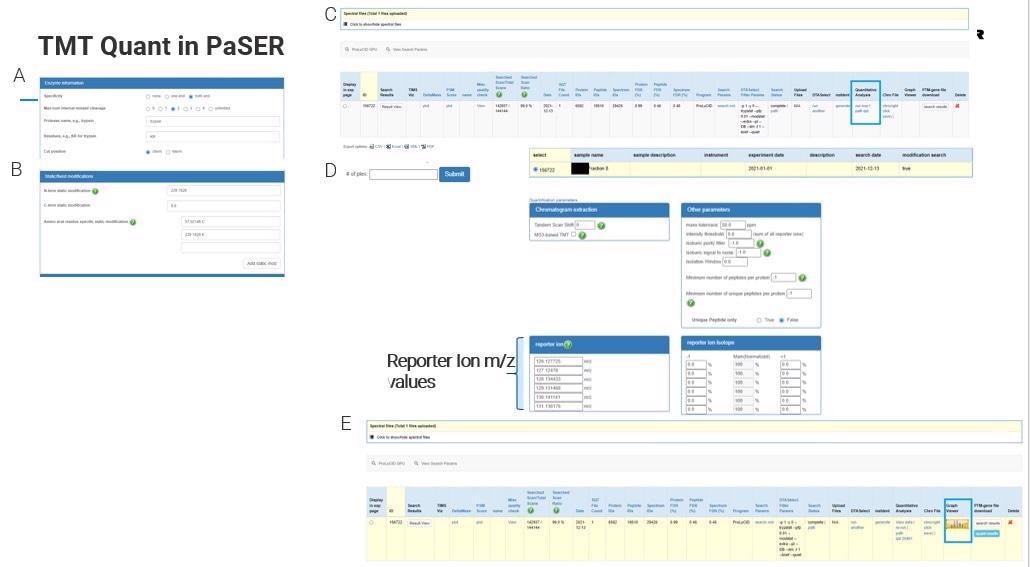
Image Credit: Bruker Daltonics
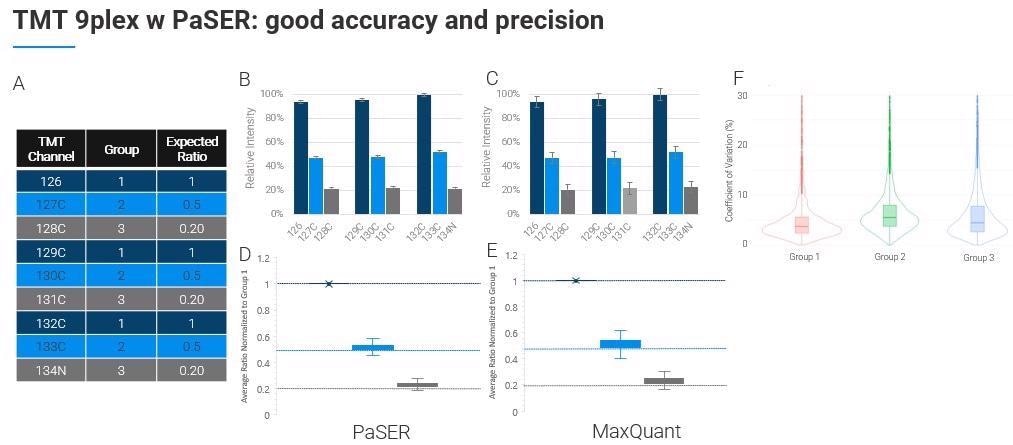
Image Credit: Bruker Daltonics
Where does the new TIMScore feature fit into this process?
TIMScore leverages the fourth dimension – the ability to predict the collisional cross section (CCS) using a machine learning model. We have been working closely with a talented grad student named Ty Garrett in Prof. John Yates lab, who has come up with a model able to accurately and reproducibly predict collisional cross-sections.
We can use these predicted collisional cross-sections and reference them against the experimentally determined collisional cross-section, calculating how well the predicted CCS fits against the experimentally determined CCS.
Where this feature becomes critical is when we need to do the discriminant analysis, choosing real from decoy hits. When we incorporate this additional dimension - TIMScore - we can look at the data from the perspective of a data cuboid. This means that instead of cutting a line, we can use a curvature fit to identify all those features that we were not previously able to find.
In practice, this allows TIMScore to increase the number of identified peptides, proteins and PSMs dramatically while maintaining the confidence level of 1% FDR.
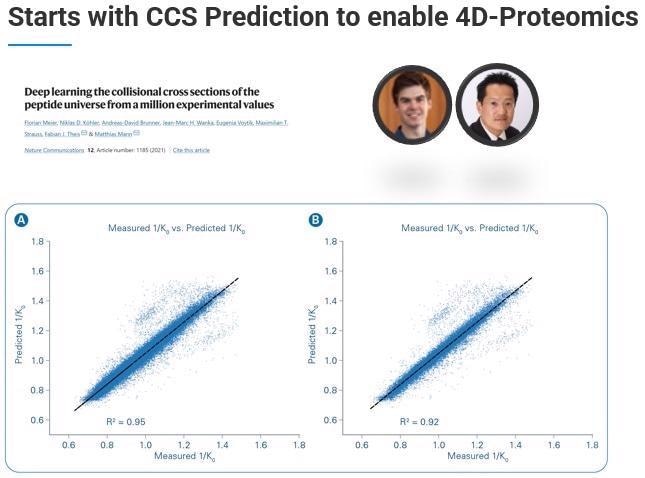
Image Credit: Bruker Daltonics
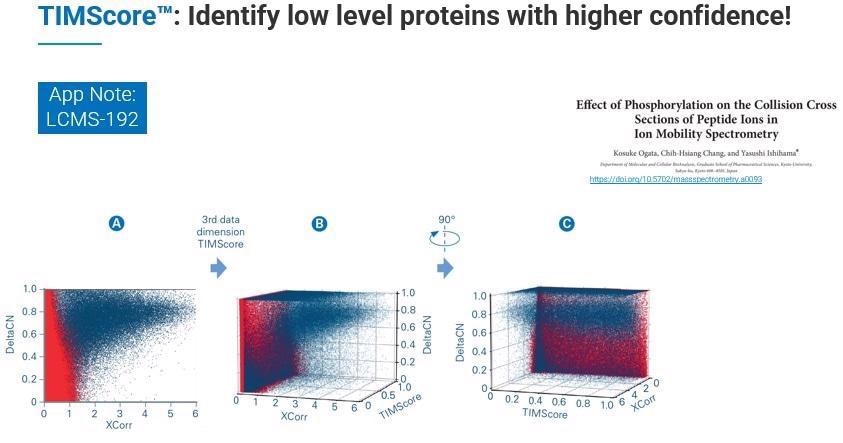
Image Credit: Bruker Daltonics
Have any studies been done to explore the potential of TIMScore in real-world applications? Can you provide examples, if so?
We tested the TIMScore in-house by taking some published literature from the lab of Yasushi Ishihama at Kyoto University that was looking at tryptic and phosphorylated peptides and comparing the results both with and without TIMScore.
We determined that by using the TIMScore, we were effectively able to double the number of peptides identified from previous reports.
Where this becomes critical is in the increased protein sequence coverage. Instead of observing 5.5 or 7.5 peptides per protein, now we are observing 15 peptides per protein.
This boost in sequence coverage is important because we are not discovering these new proteins at random – we are digging deeper into and expanding the sequence coverage of proteins we knew existed, but we just did not have the capability to detect formally.
We found that the TIMScore works exceptionally well at the phosphopeptide level. This is important because normally, when we fragment a phosphopeptide, we get a neutral loss and a weak peptide b and y-ion series, making it hard for database search algorithms to confidently detect this.
The collisional cross-section, or 4D-Proteomics, helps pick up those peptides that we were not above able to assign initially.
Dr. Stan Stevens has been working with us on this example application, and we have already seen a good increase in the number of observable phosphopeptides in his data sets.
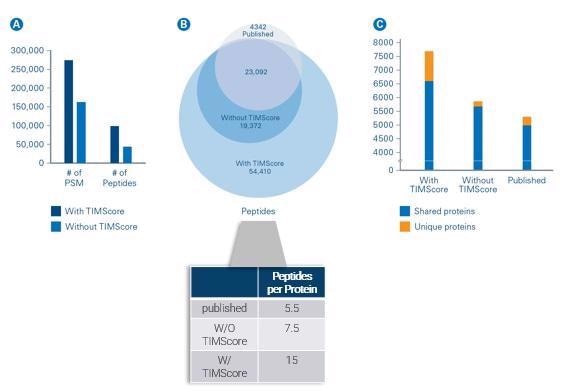
Image Credit: Bruker Daltonics
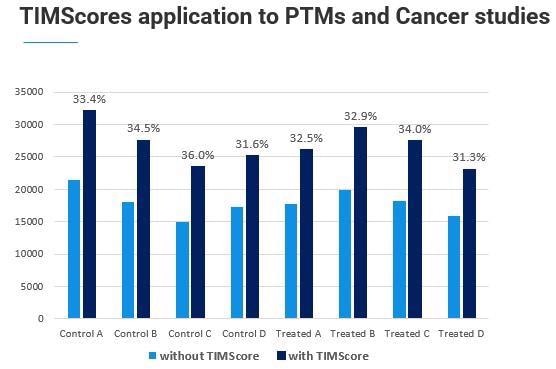
Image Credit: Bruker Daltonics
What is TIMS DIA-NN, and how can this tool be leveraged in a lab setting?
With a powerful means of performing database searches using the PaSER box and with the TIMScore generated in real-time, we wanted to address a further quantitative need.
The adoption of timsTOF technology has been strong, and notably we have seen more researchers using a dia-PASEF approach and then using algorithms like DIA-NN, Spectronaut or MaxDIA to do the analysis. The DIA-NN algorithm was developed by Drs. Vadim Demichev and Markus Ralser and has really benefited the proteomics community.
We have successfully retooled and integrated DIA-NN into a vendor specific and CCS-enabled algorithm into the PaSER box in the form of the new TIMS DIA-NN feature.
The new libraries are larger because of the additional CCS dimension and TIMScore which enable improved identification. In fact, Robin Park and team have added 20 additional features to the TIMS DIA-NN to accommodate this extra dimension.
TIMS DIA-ANN is able to provide label-free quantitation and match-between-runs (MBR) across a large sample cohort.
To look at how this works in a typical lab setting, we can use an example of the number of protein groups identified from 200 ng of K562 lysate. Over 8,300 proteins were identified using a PepSep 25 column with nanoElute on a timsTOF Pro 2 and a TIMScore powered spectral library using TIMS DIA-NN. This was done in just a short gradient of 35 minutes.
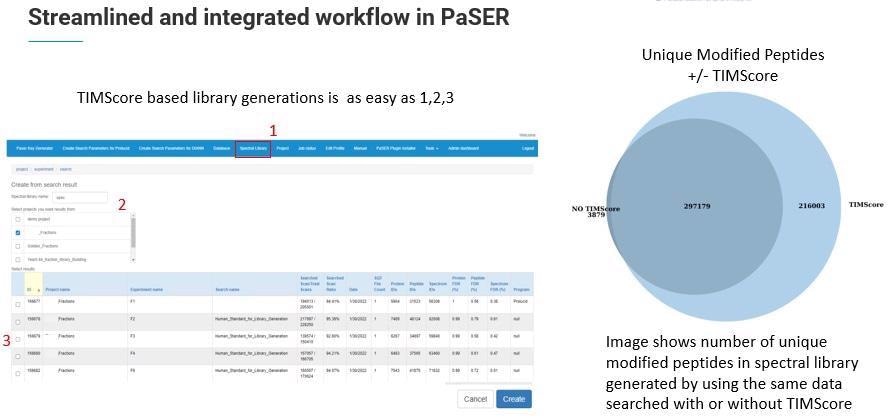
Image Credit: Bruker Daltonics
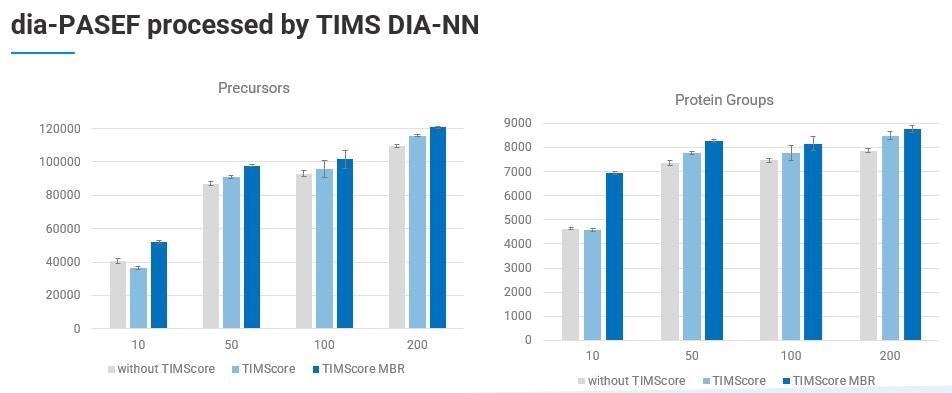
Image Credit: Bruker Daltonics
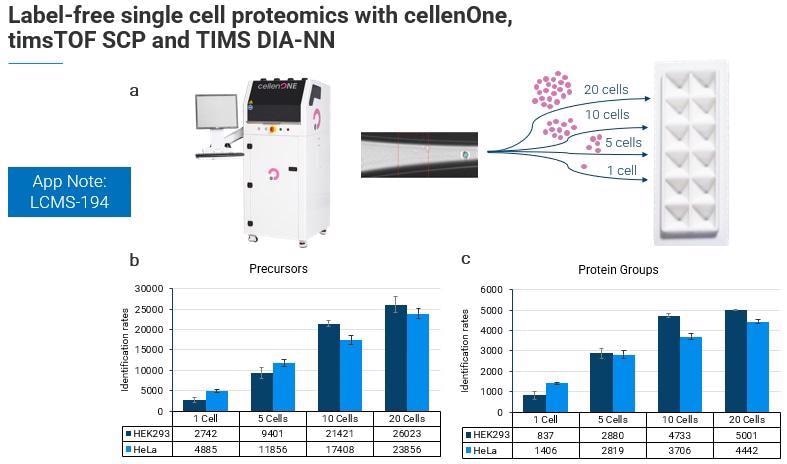
Image Credit: Bruker Daltonics
Bruker Daltonics the leader in Mass Spectrometry Solutions Explaining TIMS within timsTOF Technology
Is there anything else that you would like our readers to know?
First I should mention the PaSER’s compatibility. PaSER works with the timsTOF, the timsTOF Pro 2, the timsTOF SCP and the timsTOF FLEX - the entire timsTOF series is applicable.
We have also had some excellent results working with the cellenOne team toward single cell proteomics, where we have used a timsTOF SCP and TIMS DIA-NN to sort different numbers of cells and different cell types.
Even when working at the single-cell level with true label-free quantitation in single-cell proteomics and without a boosting channel, the number of precursors and protein groups identified is quite remarkable.
We know that HeLa cells are about six times larger than HEC 293 cells, and in this experiment, the researchers noticed that the heterogeneity of the cell types starts to become more evident as these different cell populations are pooled.
All of the examples I have shared showcase the outstanding capabilities of PaSER with timsTOF technology. PaSER provides a genuinely ‘run and done’ user experience, even when working with TMT and at single cell level.
Leveraging its powerful features expands its capabilities further, with TIMScore enabling a fourth dimension that provides more PSM, peptide and protein IDs and TIMS DIA-NN proving a new dia-PASEF workflow that is fully integrated into the PaSER box.
About Bruker Daltonics

Discover new ways to apply mass spectrometry to today’s most pressing analytical challenges. Innovations such as Trapped Ion Mobility (TIMS), smartbeam and scanning lasers for MALDI-MS Imaging that deliver true pixel fidelity, and eXtreme Resolution FTMS (XR) technology capable to reveal Isotopic Fine Structure (IFS) signatures are pushing scientific exploration to new heights. Bruker's mass spectrometry solutions enable scientists to make breakthrough discoveries and gain deeper insights.