Why are therapeutic antibodies traditionally produced in vivo during early drug discovery?
Typically, in vivo models are used because they are cost-effective and relatively fast at generating candidate antibodies that can be tested. Introduced by Köhler and Milstein in 1975, the traditional mouse hybridoma technique begins by injecting a validated or known antigen into the animal model, for example, mice, which causes an immune response that generates novel antibodies. These novel antibodies can then be harvested and fully characterized downstream to ensure that they have the desired molecular function and properties.
![]()
Image credit: extender_01 | Shutterstock
Scientific and technological advances have further improved in vivo studies. More recently, humanized mice have been immunized to express human immunoglobulins, rather than the mouse antibody genes, which enhances how translational these are downstream.
Why are scientists moving away from producing antibodies within animal models?
Animal models continue to be prevalent, primarily because they are tried and tested in the industry with good results. Despite the challenges, hybridoma technology is a well-established platform, and the humanization of animal models has further promoted this method. Most FDA approved therapeutics have been produced using humanized platforms.
However, fully rodent animal models can result in rodent specific immunogenicity and proteins from the animal model may remain at a later stage. In fact, not all antibodies produced in humanized animal models will be entirely human.
Other technologies, for instance, phage display provide a compelling alternative as they don’t require animal models, have shorter timelines and researchers can use a fully human antibody library. These libraries can be quite diverse and can contain different constructs within them, for example, single-domain antibodies. Using next generation sequencing (NGS) with phage display helps characterize immunoglobulin genes from large populations, which better represents natural human antibodies.
Phage display isn’t without downsides either; as it is a prokaryotic system rather than mammalian, there may be issues with affinity and other properties of this in vitro platform.
What characteristics are scientists looking for when they analyze entire populations of antibody-producing B-cells?
After successfully immunizing an animal, a lymphoid organ, for example, the spleen, is harvested. This is processed and B-cells are isolated. B-cells that produce antigen-specific antibodies that bind to the target antigen then need to be separated from those that bind to other antigens.
Next, the reactivity to the target antigen is tested to ensure that selected antibodies bind specifically to the antigen (rather than to a range of antigens) and that this affinity and sensitivity is above the required level. Once the unique B-cells have been isolated, scientists will screen for binding characteristics, cross-reactivity and more.
What techniques are commonly used to screen B-cells for these characteristics?
Research teams will typically use flow cytometry to screen the B-cells directly or more often produce hybridomas which are then screened.
Producing a hybridoma population requires a fusion reaction between the B-cell and a cancerous myeloma cell. This allows the B-cell to proliferate and survive indefinitely in culture, which is essential to safeguard potential cells of interest. Unfortunately, when creating hybridomas, the whole B-cell population is unable to be fused; therefore, only a fraction of the total population can be screened, thus leading to the loss of a large part of the B-cell repertoire.
Traditionally, flow cytometry is favored because of its high-throughput, and antibodies secreted from the B-cells can potentially be screened using cold capture, a technique used to manipulate the cell to prevent the full secretion of antibodies by trapping them at the cell surface. However, this is a representation rather than direct measurement of antibody secretion.
Alternative screening methods include ELISA and Elispot, which can measure B-cell secretion. Although these techniques often need to be completed manually, making them tedious and limited to low-scale screening.
These techniques can create a harsh environment for high-value cells. How does this affect cell viability and therefore, productivity?
A research team typically spends many months during this process cycle, from immunizing the animal model, collecting cells and creating hybridomas or directly screening B-cells. The population of target cells can be as low as 0.001% of the original ~40 million cells in the harvested organ. Further loss due to diminishing cell viability and cell death due to the harsh processing of these cells can severely impact productivity.
After completing this cycle, the target cells may not be found because they have either not been successfully fused during the hybridoma fusion process or died during culture due to the harsh processing. This is a big challenge in the industry.
How does the single cell analysis system that you developed overcome these issues?
The Cyto-Mine® Single Cell Analysis System can overcome numerous challenges in the industry. First, the need for high-throughput yet gentle technology that enables the deep interrogation of whole B-cell repertoires and hybridoma populations while preserving cell viability. Second, the need for highly sensitive and specific assays to find rare antibody-secreting cells with the desired antibody characteristics.
Cyto-Mine® Operations Guide
Cyto-Mine® can be used in screening entire cell populations to find that rare B-cell or hybridoma that secretes the antigen-specific antibody. In fact, researchers can input isolated B-cells directly into the Cyto-Mine®, which drastically cuts down the process cycle timelines, and removes the possibility of losing cells in culture. Additionally, Cyto-Mine® offers the flexibility in assay design, of which there is a great need due to the varying nature of the targets and the biologics.
Underpinned by Sphere Bio's patented picodroplet technology, Cyto-Mine® integrates the selective screening of tens of millions of single cells in miniaturized aqueous picoliter compartments called picodroplets.
Picodroplets provide individual micro-reaction chambers where secreted proteins accumulate and can be assayed to identify valuable proteins (e.g. antibodies with antigen-specificity), to find rare cells of interest or to find high-producing cells.
The picodroplets themselves provide a protective environment for the fragile B-cells, shielding them from shear stress as the picodroplets travel through the microfluidic channels. Cells can also be encapsulated in their preferred culture media, so there are no harmful culture transitions.
By combining B-cell isolation and identification of ‘antigen-specific hits’ into a process that takes only 1-2 days in Cyto-Mine®, you significantly reduce the stress on the fragile cells and accelerate the steps to downstream analysis such as functional testing or sequencing.
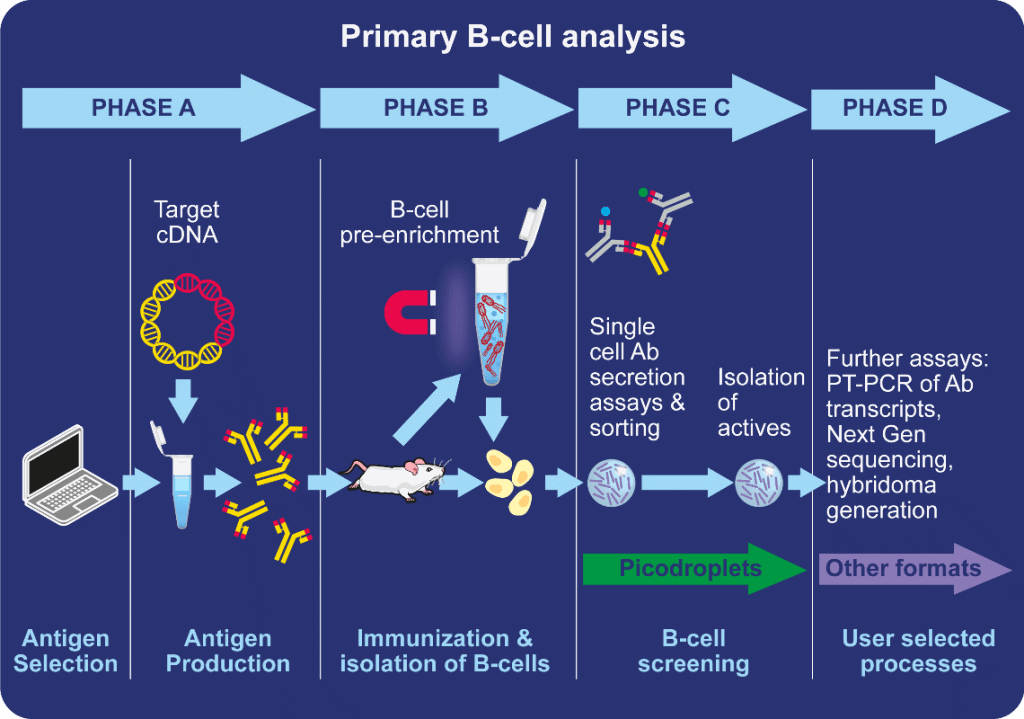
Image credit: Sphere Bio
How does the technology determine antigen-specificity?
Users must know whether the antibody is an IgG or an IgM, before making detection probes that will bind to the target of interest. The assay format is based on a simple FRET reaction, and two detection probes must be encapsulated into the picodroplet along with the cells. One that binds to the Fc region of the antibody and another that targets the antigen on that antibody. Both probes are conjugated to a fluorophore, making a pair of detection probes that can induce a FRET reaction.
When the detection probes are in close proximity, i.e. if they have bound to the Fc and Fab regions of the secreted antibody, they will induce a FRET-mediated shift on fluorescence where the excitation of one probe will transfer energy to the other which will emit a fluorescent signal. Cyto-Mine® detects for an energy transfer within the picodroplet, and as the detection probes are customized to target only the antigen of interest, there will not be a fluorescent energy transfer if the antigen of interest isn’t present.
Cyto-Mine® supports flexible assay design, so the assay format can be tailored to make it specific to the target.
Was automation important for you when developing this system? If so, why?
Developed with biopharma in mind, the initial design of Cyto-Mine® relied on input from key pharma companies, to solve their most significant challenges and automation was vital to do this.
The current process is incredibly time-consuming and tedious. New techniques need to have a high-throughput, diversity and flexibility of the assay. Rather than automating pipetting, we’ve made an entirely novel system to approach antibody discovery.
The Cyto-Mine® is also applicable to cell line development, where there is a similar need to automate inefficient processes, and Cyto-Mine® has single cell imaging and dispensing capacity making it a great fit.
How do you think the process of antibody discovery will evolve over the next decade? What impact will this have on the pharmaceutical industry and the therapies that are available for patients?
There’s an incredibly high demand for antibody-based biotherapeutics, and it’s easy to see how successful they are with over 500 antibody-based therapeutics beginning to come through clinical trials. This demand is exemplified with one of the Antibody Society’s 20 antibodies to watch in 2020, an antibody against the Ebola virus. The company producing this antibody could stop trials before completion because it was so successful compared to any other treatments.
Over the next decade, the antibody discovery process needs to become quicker, and as new technology is incorporated, should become more rapid. This optimization will shorten the time to market and get new therapeutics in the hands of patients faster.
Where can our readers find more information?
About Fluidic Sciences and Sphere Bio
Fluidic Sciences develops transformative in‑solution technologies for protein interaction analysis. Its flagship Fluidity One‑M instrument leverages Microfluidic Diffusional Sizing (MDS) to measure binding affinity, stoichiometry, size, and concentration without immobilization - directly in complex backgrounds such as serum, plasma, and lysate.
Sphere Bio is a brand of Fluidic Sciences. Its technology develops and manufactures single‑cell analysis and monoclonality assurance systems that enable researchers to find, analyze, and isolate the most valuable cells with speed and precision. Its proprietary picodroplet microfluidics and Cyto‑Mine® Chroma multiplexing platform power applications across antibody discovery, cell line development, cell engineering, and cell therapy.