A recent study reveals how the non-structural protein 1 (NSP1) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) plays a crucial role in inhibiting the host cell protein synthesis but simultaneously facilitating the viral protein synthesis.
The study by researchers from Harvard Medical School, Boston Children's Hospital, Ohio State University, USA, and Harbin Institute of Technology, China, is currently available on the bioRxiv* preprint server.
SARS-CoV-2, the causative pathogen of coronavirus disease 2019 (COVID-19), is an enveloped, single-stranded RNA virus with a genome size of approximately 30 kb. After infecting host cells, SARS-CoV-2 synthesizes a wide range of structural and non-structural proteins using the host cell protein synthesis machinery. While the structural proteins of SARS-CoV-2, such as spike protein is essential for viral entry into host cells, non-structural proteins, particularly NSP1, plays a crucial role in suppressing the host cell protein expression, which in turn facilitates viral replication and evasion of host immune responses.
Regarding host cell protein synthesis, it is known that the C-terminal domain of NSP1 directly binds to 40S small ribosomal subunit to inhibit host mRNA translation. However, it is still unknown how SARS-CoV-2 maintains its own protein synthesis by bypassing NSP1-mediated inhibition of host protein synthesis machineries. The current study has been undertaken to address this vital question.
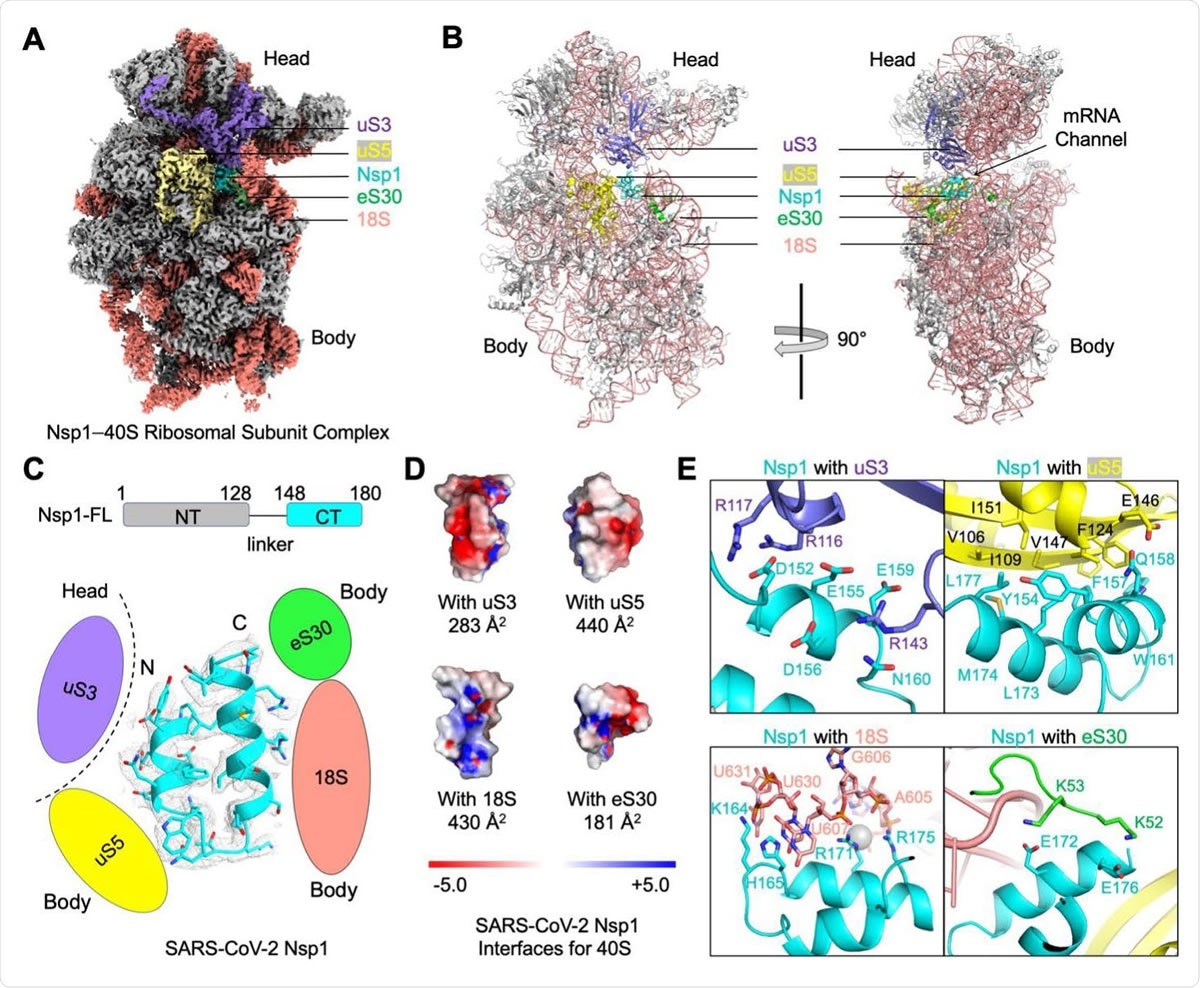
Blockage of the mRNA Channel in the 40S by the Nsp1 C-terminal Helical Hairpin (A) Cryo-EM density of the Nsp1-40S complex. Nsp1 is in cyan. Subunits of 40S that interact with Nsp1 are color-coded. (B) Ribbon diagram of the Nsp1-40S complex. Nsp1 (cyan) binds at the mRNA channel in the cleft between the head and body of the 40S ribosomal subunit. Subunits of 40S that interact with Nsp1 are color-coded. (C) Ribbon diagram and cryo-EM density of SARS-CoV-2 Nsp1 with schematics highlighting its interfaces to 40S. (D) Electrostatic surface representations of Nsp1 binding surfaces to the 40S subunits and rRNA. Buried surface areas are marked. (E) Detailed interactions between Nsp1 and the 40S ribosomal subunit.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Current study design
Regarding other coronaviruses, such as SARS-CoV, it has been shown that the 5' untranslated region (UTR) of the viral mRNA plays an important role in escaping NSP1-mediated suppression of mRNA translation. Given this observation, the current study scientists aimed at investigating how NSP1 of SARS-CoV-2 hijacks host protein synthesis machineries to facilitate viral mRNA translation while inhibiting host mRNA translation.
Important observations
In agreement with previous study findings, the current study revealed that the interaction between the helical hairpin at the C-terminal domain of viral NSP1 and the eukaryotic 40S small ribosomal subunit prevents the entry of mRNA into the host ribosome, and thus, inhibits the host protein synthesis. However, they observed that in the case of SARS-CoV-2, the N-terminal domain of NSP1 directly binds to the 5' UTR of viral mRNA, leading to the release of translational inhibition imposed by NSP1 and efficacious synthesis of viral proteins. Another important phenomenon they observed was that covalent linking between the C-terminal and N-terminal domains of NSP1 is required for the evasion of NSP1-mediated translational suppression.
Furthermore, the scientists observed that the binding of 5' UTR of SARS-CoV-2 mRNA with the N-terminal domain of NSP1 prevents the interaction between the NSP1's C-terminal domain and 40S ribosomal subunit, probably because of steric interference, which prevents the C-terminal domain from reaching the 40S subunit. As a result, viral mRNA could enter the ribosome to be translated into proteins. Moreover, they revealed that among four stem-loop structures of 5' UTR of SARS-CoV-2 mRNA, stem-loop1 (SL1) plays a crucial role in escaping NSP1-mediated translational inhibition.
To further support their findings, the scientists increased the length of the linker between the C-terminal and N-terminal domains of NSP1, which led to a significant reduction in viral mRNA translation. With this finding, the scientists hypothesized that even if NSP1 is already bound to the 40S head region, the interaction between 5' UTR of viral mRNA and N-terminal domain of NSP1 is capable of dislodging the C-terminal domain of NSP1 from the 40S ribosomal subunit. One possible reason for this is that because of the short linker length; a pulling force is generated upon binding of 5' UTR of SARS-CoV-2 mRNA and N-terminal domain of NSP1, which subsequently dislodges the C-terminal domain of NSP1 from the 40S ribosomal subunit.
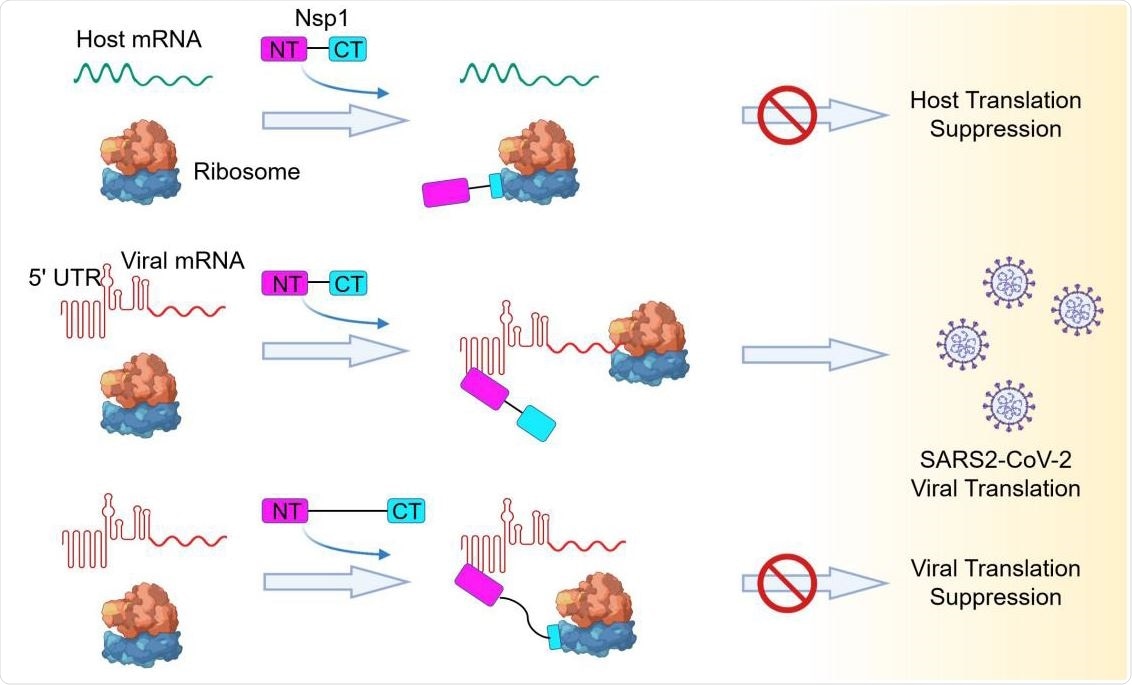
Model of a Bipartite Mechanism for Nsp1-Mediated Translation Inhibition and Evasion by SARS-CoV-2 5’ UTR A schematic model depicting the bipartite roles of SARS-CoV-2 Nsp1 during infection. First, Nsp1 blocks host mRNA from binding to the 40S ribosomal subunit due to physical occlusion by the bound Nsp1-CT. Second, Nsp1 supports viral mRNA translation by interacting with SARS-CoV- 2 5’ UTR using Nsp1-NT, which results in dissociation of the Nsp1-CT-40S complex to overcome inhibition. This mechanism of evasion of Nsp1-mediated translation inhibition is illustrated by the failure of linker-lengthened Nsp1 to support viral mRNA translation. With the longer linker, the Nsp1-NT-5’ UTR complex can co-exist with the Nsp1-CT-40S complex.
Study significance
The current study identifies a novel mechanism by which SARS-CoV-2 overcomes the translational inhibition imposed by NSP1 and utilizes the host protein synthesis machineries to translate its own mRNA. Given the important function of stem-loop1 in facilitating viral protein synthesis, it can be utilized as a potential therapeutic target to inhibit SARS-CoV-2 replication. Moreover, the growth and survival of SARS-CoV-2 inside host cells can be prevented by inhibiting the interaction between 5' UTR of SARS-CoV-2 and N-terminal domain of NSP1 using small molecule inhibitors or antisense RNAs.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.