Scientists worldwide are fighting to contain the ongoing COVID-19 pandemic, which has already claimed more than 2.48 million lives. Preventing spillback infections is essential to containing the pandemic. To do so, it is vital to understand the nature of the virus, i.e., its susceptibility to a range of hosts.
Like previous disease outbreak episodes, SARS-CoV-2 originated from the spillover of a zoonotic pathogen. This virus's high transmission rate is attributed to its capability to use a highly conserved cell surface receptor, angiotensin-converting enzyme 2 (ACE2), to enter the host cell.
The ACE2 protein is predominantly present on the cell surface of all major vertebrate groups. The high rate of spillback infections in the past year can be explained by the simultaneous occurrence of three factors – (a) a high human population, (b) high infection potential of SARS-CoV-2, and (c) the ubiquitous nature of the ACE2 receptor.
Now, a new study, published on the bioRxiv* preprint server, has revealed the importance of spillback infections during pandemics. During spillback infections, the SARS-CoV-2 virus was observed to have been transmitted from humans (host) to other mammals. Such spillback infections may lead to the establishment of a new host (animals), which may act as a virus reservoir. These animals may, subsequently, cause a secondary spillover to humans.
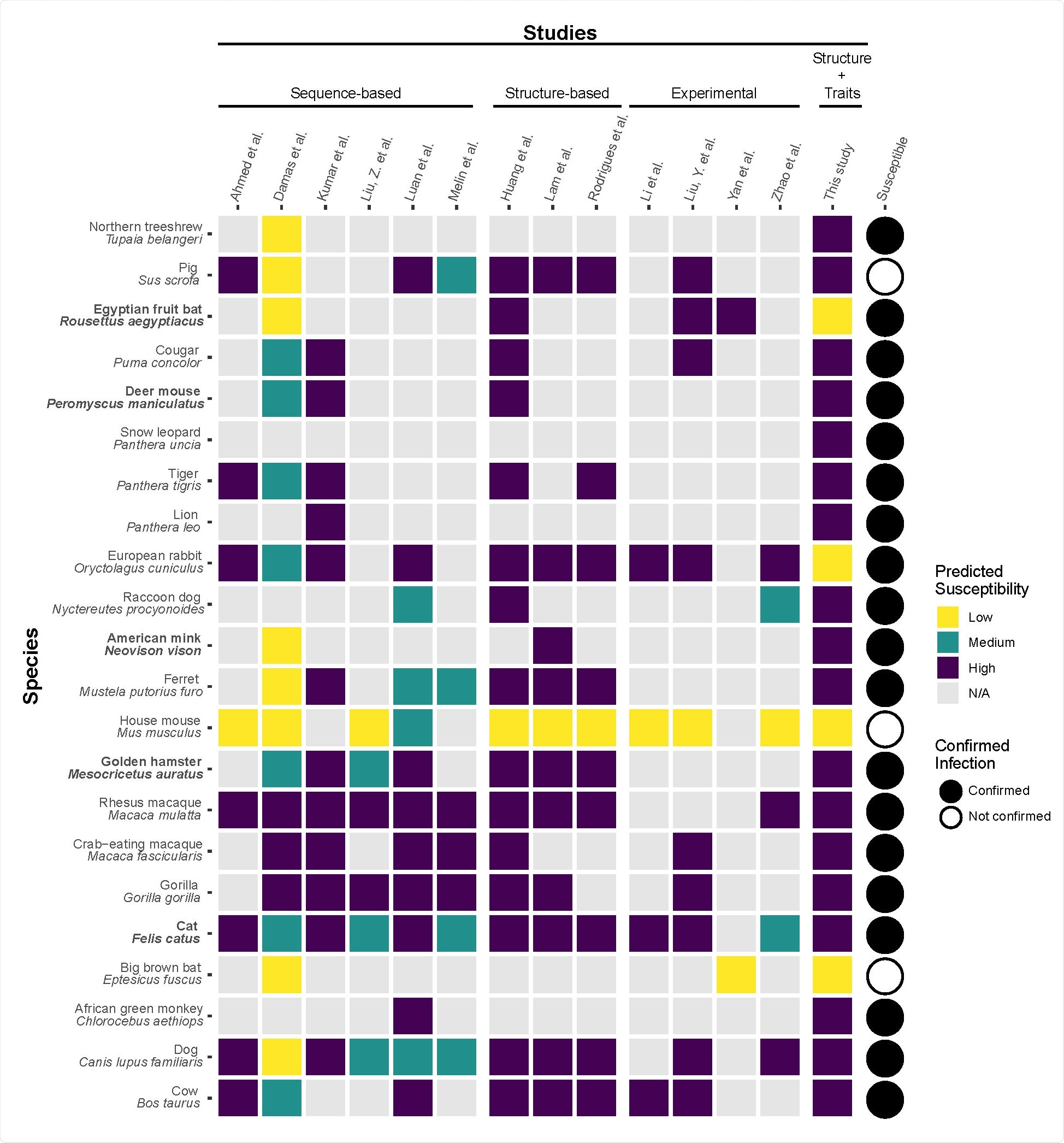
A heatmap summarizing predicted susceptibility to SARS-CoV-2 for species with confirmed infection status from in vivo experimental studies or documented natural infections. Studies that make predictions about species susceptibility are shown in the x-axis, organized by method of prediction (those relying on ACE2 sequences, estimating binding strength using three dimensional structures, or laboratory experiments). Predictions about zoonotic capacity from this study are listed in the second to last column, with high and low categories determined by zoonotic capacity observed in Felis catus. Confirmed infections for species along the y-axis are summarized in (Gryseels et al., 2020) and are depicted as a series of filled or unfilled circles. Bolded species have been experimentally confirmed to transmit SARS105 CoV-2 to naive conspecifics. Species predictions ranged from warmer colors (yellow: low susceptibility or zoonotic capacity for SARS-CoV-2) to cooler colors (purple: high susceptibility or zoonotic capacity). See supplementary file 1 for detailed methods about study categorization.
Such an occurrence was found in Denmark and in the Netherlands, where SARS-CoV-2 was transmitted from humans to farmed mink, and afterward, a variant of the virus was transmitted back from the mink to humans.
The secondary spillover poses a significant threat as it generates mutant strains. A variety of pets, farmed animals, etc., are reported as new hosts of SARS-CoV-2. The emergent strains may be more virulent than the existing ones. A prior study of the recent spillover event has revealed that the mink-derived variant can decrease sensitivity to neutralizing antibodies. This indicates that the efficacy of vaccines may also take a toll.
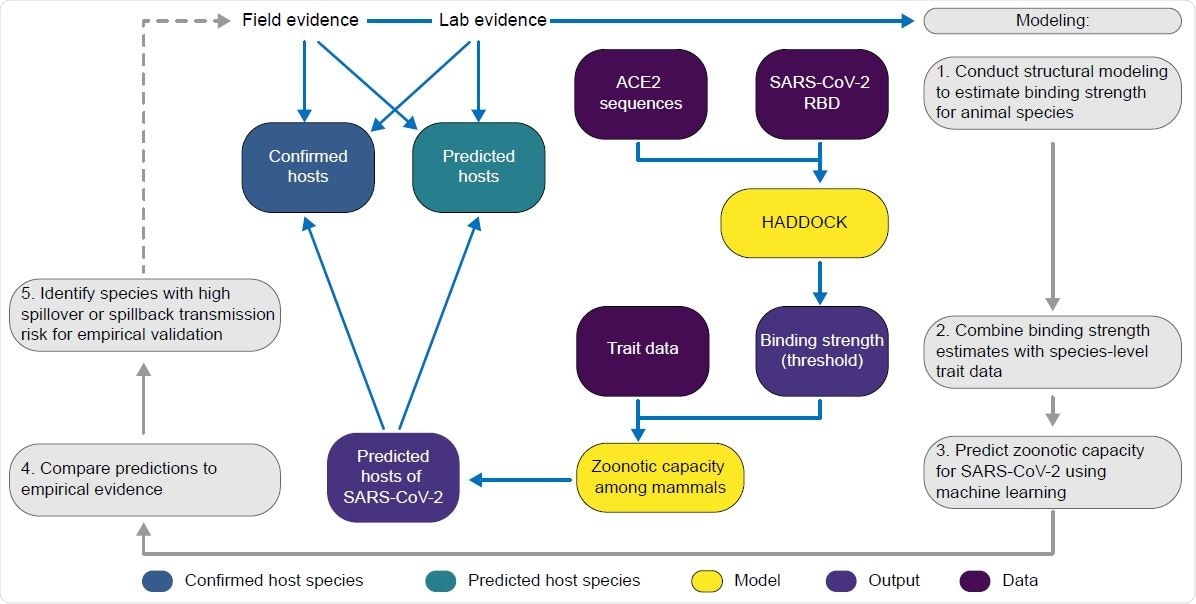
A flowchart showing the progression of our workflow combining evidence from limited lab and field studies with additional data types to predict zoonotic capacity across mammals through multi-scale statistical modeling (gray boxes, steps 1-5). For all vertebrates with published ACE2 sequences, we modelled the interface of species' ACE2 bound to the viral receptor-binding domain using HADDOCK. We then combined the HADDOCK scores, which approximate binding strength, with species’ trait data and trained machine learning models for both mammals and vertebrates (yellow boxes). Mammal species predicted to have high zoonotic capacity were then compared to results of in vivo experiments and in silico studies that applied various computational approaches. We then identified a subset of species with a particularly high risk of spillback and secondary spillover potential to prioritize additional lab validation and field surveillance (dashed line).
Scientists have carried out various computational studies to make valid predictions about non-human animals susceptible to SARS-CoV-2. To this end, both comparative models of ACE2 orthologs sequences among various species and structure-based models of the viral spike protein bound to ACE2 orthologs were used.
The current study develops a new model by combining structure-based inference with species-level trait data. This would help predict the zoonotic capacity of different animal species in becoming zoonotic hosts of SARS CoV-2.
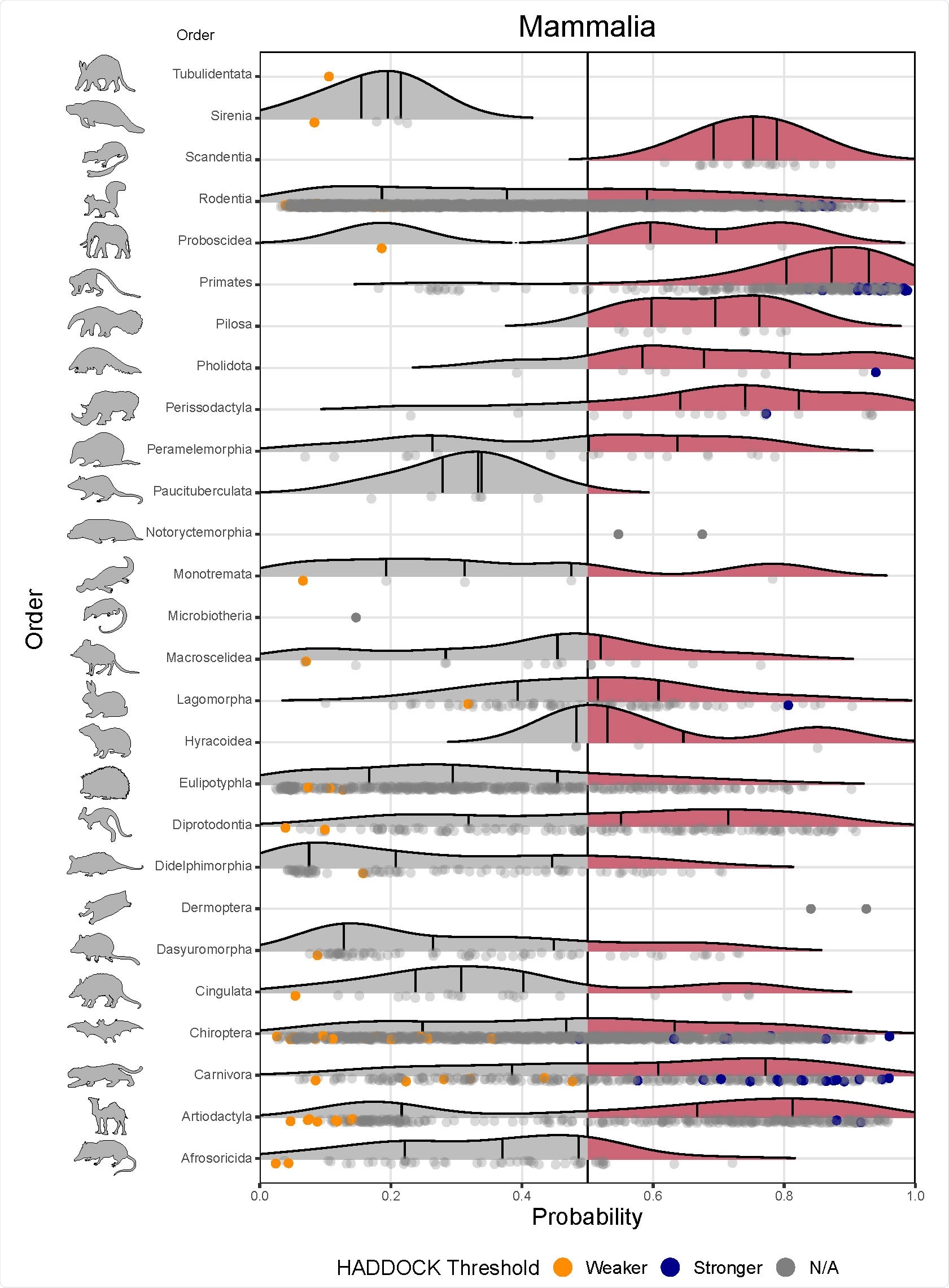
Ridgeline plots showing the distribution of predicted zoonotic capacity across mammals. 379 Predicted probabilities for zoonotic capacity across the x-axis range from 0 (likely not susceptible) to 1 380 (zoonotic capacity predicted to be the same or greater than Felis catus), with the vertical line representing 381 0.5. The y-axis depicts all mammalian orders represented by our predictions. Density curves represent 382 the distribution of the predictions, with those parts of the curve over 0.5 colored pink and lines 383 representing distribution quartiles. The predicted values for each order are shown as points below the 384 density curves. Points that were used to train the model are colored: orange represents species with weaker predicted binding, blue represents species with stronger predicted binding. Selected family-level 386 distributions are shown in the two figure supplements for this figure.
To enhance the predictive capacity of the models across species, scientists have incorporated intrinsic biological traits of ~5400 mammal species in their study.
In the course of model development, several factors that are associated with species susceptibility to SARS-CoV-2 were considered, for example, the binding strength between SARS-CoV-2 receptor binding domain and host ACE2. The combined modeling system has helped increase the accuracy percentage of prediction of zoonotic capacity to 72%.
The model has predicted a high SARS-CoV-2 zoonotic capacity for many domesticated, farmed, and live traded animal species. For example, among all livestock, Bubalus bubalis (water buffalo) was predicted to have a high zoonotic capacity. Some of the other species that showed a high percentage of zoonotic capacity are rats, rodents, and bats. Endangered species such as mountain gorillas and addax also have a high zoonotic capacity, thereby putting the individuals directly involved in active conservation management at greater risk of spillback transmission. The model has also helped to identify various other animal species that can act as hosts to SARS-CoV-2.
Despite a high level of agreement between these models and empirical studies' predictions, some differences remain between the computational results and the actual experiments, including animals. As an example, several computational predictions suggested that Sus scrofa (pigs) show susceptibility to SARS-CoV-2, but this result did not hold up in animal-based experiments.
Scientists believe that the disagreement between the real-world observations and in silico predictions of zoonotic capacity stems from the fact that the host susceptibility and transmission capacity are not the sole determinants. Transmission of the virus also depends upon the host's cellular environments, i.e., host immunogenicity, protein receptors, etc., where the virus's replication takes place. Further, other challenges such as limitations in the ACE2 sequences and species trait data also aid in predicting error.
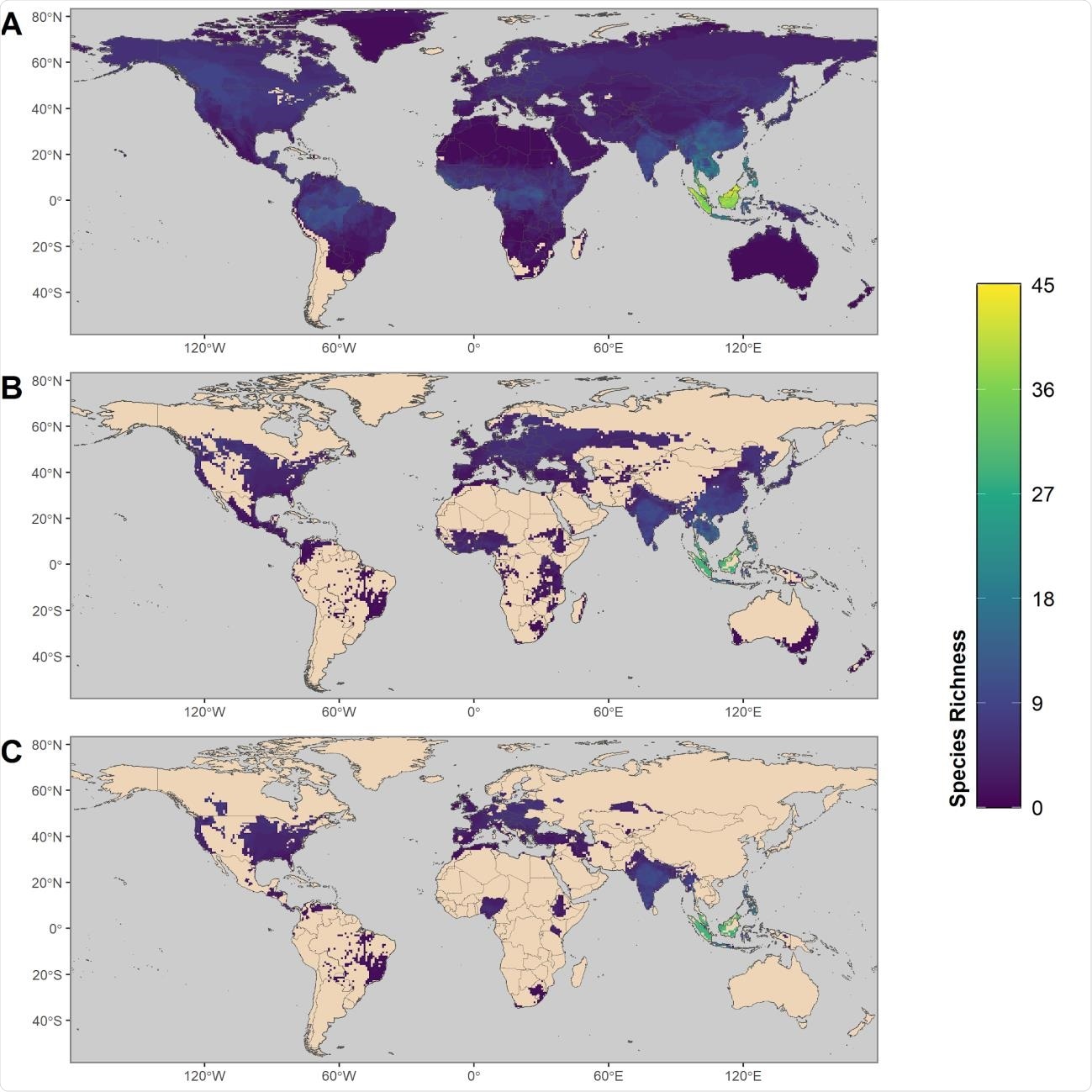
Maps showing the global distribution of species with the predicted capacity to transmit SARS-CoV452 2. (A) depicts global species richness of the top 10 percent of model-predicted zoonotic capacity. Ranges of this subset of species were filtered to those associated with human-dominated or human-altered habitats (B), and further filtered to show the subset of species that overlaps with areas of high human SARS-CoV-2 positive case counts (over 100,000 cumulative cases as of 15 February 2021) (C)
Therefore, researchers have recommended that a single methodology is not sufficient to predict the zoonotic capacity of SARS-CoV-2. The zoonotic predictive capacity could be increased by assessing the results obtained from combining theoretical models, statistical models, laboratory experiments, and real-world observations.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Predicting the zoonotic capacity of mammal species for SARS-CoV-2 Ilya R. Fischhoff, Adrian A. Castellanos, João P.G.L.M. Rodrigues, Arvind Varsani, Barbara A. Han bioRxiv 2021.02.18.431844; doi: https://doi.org/10.1101/2021.02.18.431844, https://www.biorxiv.org/content/10.1101/2021.02.18.431844v1
- Peer reviewed and published scientific report.
Fischhoff, Ilya R., Adrian A. Castellanos, João P. G. L. M. Rodrigues, Arvind Varsani, and Barbara A. Han. 2021. “Predicting the Zoonotic Capacity of Mammals to Transmit SARS-CoV-2.” Proceedings of the Royal Society B: Biological Sciences 288 (1963). https://doi.org/10.1098/rspb.2021.1651. https://royalsocietypublishing.org/doi/10.1098/rspb.2021.1651.