Certain protein modifications promote protein stability within a cell, while others promote protein degradation. Arginylation is a protein modification process that is associated with cellular stress and causes cell death or growth arrest. The role of arginylation in infectious disease is unknown.
 Study: Protein arginylation is regulated during SARS-CoV-2 infection. Image Credit: WildMedia / Shutterstock.com
Study: Protein arginylation is regulated during SARS-CoV-2 infection. Image Credit: WildMedia / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Endoplasmic reticulum stress
SARS-CoV-2 proteins use the host cell's endoplasmic reticulum (ER) for the formation of double-membrane vesicles for ribonucleic acid (RNA) synthesis. The assembly of viral particles then occurs in the ER-Golgi intermediate compartment by the structural proteins.
There is extensive use of the ER during viral replication, which leads to stress in this cellular compartment. This results in the accumulation of misfolded proteins and activation of the unfolded protein response (UPR). The UPR pathway regulates cellular events like growth, defense, homeostasis, and cell survival. This pathway activation may also boost tissue repair processes.
SARS-CoV-2 infection induces ER stress, which may be associated with patient survival.
Protein arginylation
Ubiquitination is a protein modification process that marks the protein for degradation. N-terminal arginylation of proteins is an inducer of ubiquitination and protein degradation by the N-end rule pathway.
Protein arginylation has been associated with cellular stress conditions. Under stress conditions, arginylation promotes cell death or growth arrest. ArginyltRNA-protein transferase (ATE1) is the enzyme that promotes arginylation. ATE1 is necessary to decrease mutation events when a cell is subjected to stressful conditions that damage deoxyribonucleic acid (DNA).
Methods
In the current study, the scientists examined the modulation of the N-end rule pathway and protein arginylation in Vero CCL-81 and Calu-3 cells infected after 2 hours (h), 6 h, 12 h, 24 h, and 48 h. The Vero CCL-81 cell line is derived from kidney epithelial cells extracted from an African green monkey, whereas Calu-3 is a human lung epithelial cell line.
The authors of the current study performed an in silico reanalysis of in vivo and in vitro public omics data to verify the abundance of proteins that make up the N-end rule pathway in non-infected and SARS-CoV-2 infected groups or other viral infections. This was combined with western blotting to measure the levels of arginyl-transfer RNA (tRNA)-protein transferase (ATE1) and arginylated proteins.
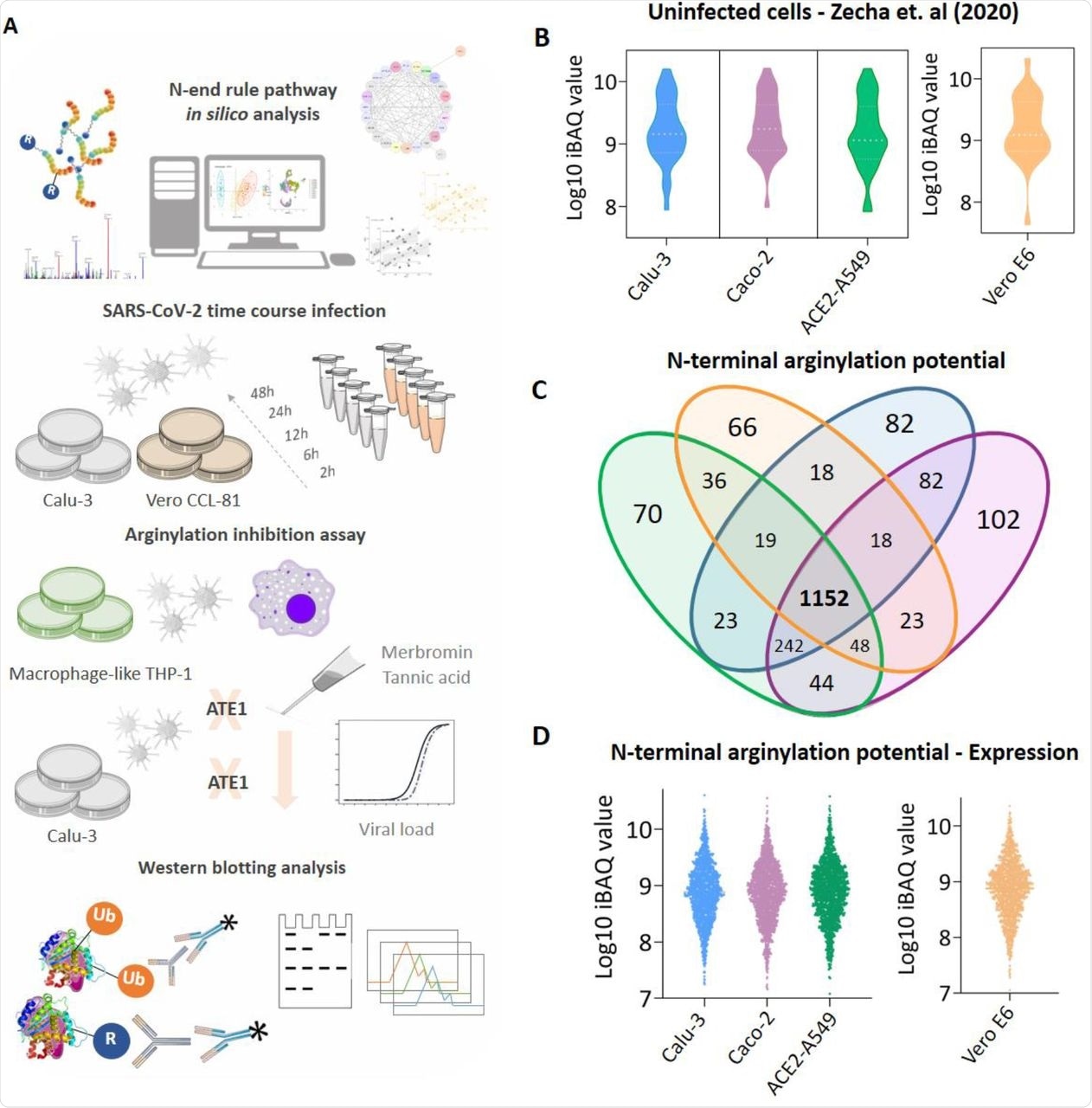 (A) Experimental workflow adopted to identify the modulation of protein arginylation during SARS-CoV-2 infection; (B) Expression profile of enzymes participating in the N-end rule pathway identified in uninfected Calu-3 (blue), Caco-2 (purple), ACE2-A549 (green), and Vero E6 (orange) cells; (C) Proteins with potential to be arginylated at N-terminal (NtE, NtD, NtC, NtN, NtQ) identified and quantified in uninfected cell models; (D) Expression profile of the 1,152 proteins with potential to be arginylated identified in uninfected Calu-3, Caco-2, ACE2-A549, and Vero E6 cells. Proteins with the potential to be arginylated were determined based on their sequences deposited in Uniprot (Homo sapiens and Chlorocebus sabaeus).
(A) Experimental workflow adopted to identify the modulation of protein arginylation during SARS-CoV-2 infection; (B) Expression profile of enzymes participating in the N-end rule pathway identified in uninfected Calu-3 (blue), Caco-2 (purple), ACE2-A549 (green), and Vero E6 (orange) cells; (C) Proteins with potential to be arginylated at N-terminal (NtE, NtD, NtC, NtN, NtQ) identified and quantified in uninfected cell models; (D) Expression profile of the 1,152 proteins with potential to be arginylated identified in uninfected Calu-3, Caco-2, ACE2-A549, and Vero E6 cells. Proteins with the potential to be arginylated were determined based on their sequences deposited in Uniprot (Homo sapiens and Chlorocebus sabaeus).
Results
Enzymes involved in protein arginylation, ubiquitination, arginine-tRNA ligase assembly, deamidation, and N-terminal methionine removal were identified in all cell models. These findings indicated that enzymes involved in the protein arginylation pathway were not modulated based on the cell type or species in uninfected conditions.
A total of 1,152 proteins can be potentially arginylated at the N-terminus. ATE1 expression was higher during infection at both transcript and protein levels. SARS-CoV-2 infected Calu-3 and Vero CCL-81 cells had statistically higher ATE1 levels as compared to the uninfected group. Thus, SARS-CoV-2 infection modulated the N-end rule pathway and increased ATE1 enzyme expression.
The scientists then verified which proteins correlated with ATE1 and analyzed their molecular function. Events related to ER and viral infection were enriched, such as protein target to ER, protein localization to ER, viral gene expression, and viral transcription. Pathways related to alterations in processes linked to RNA and coronavirus infection were also enriched. Therefore, increased ATE1 expression in SARS-CoV-2 infection correlated with events linked to the ER.
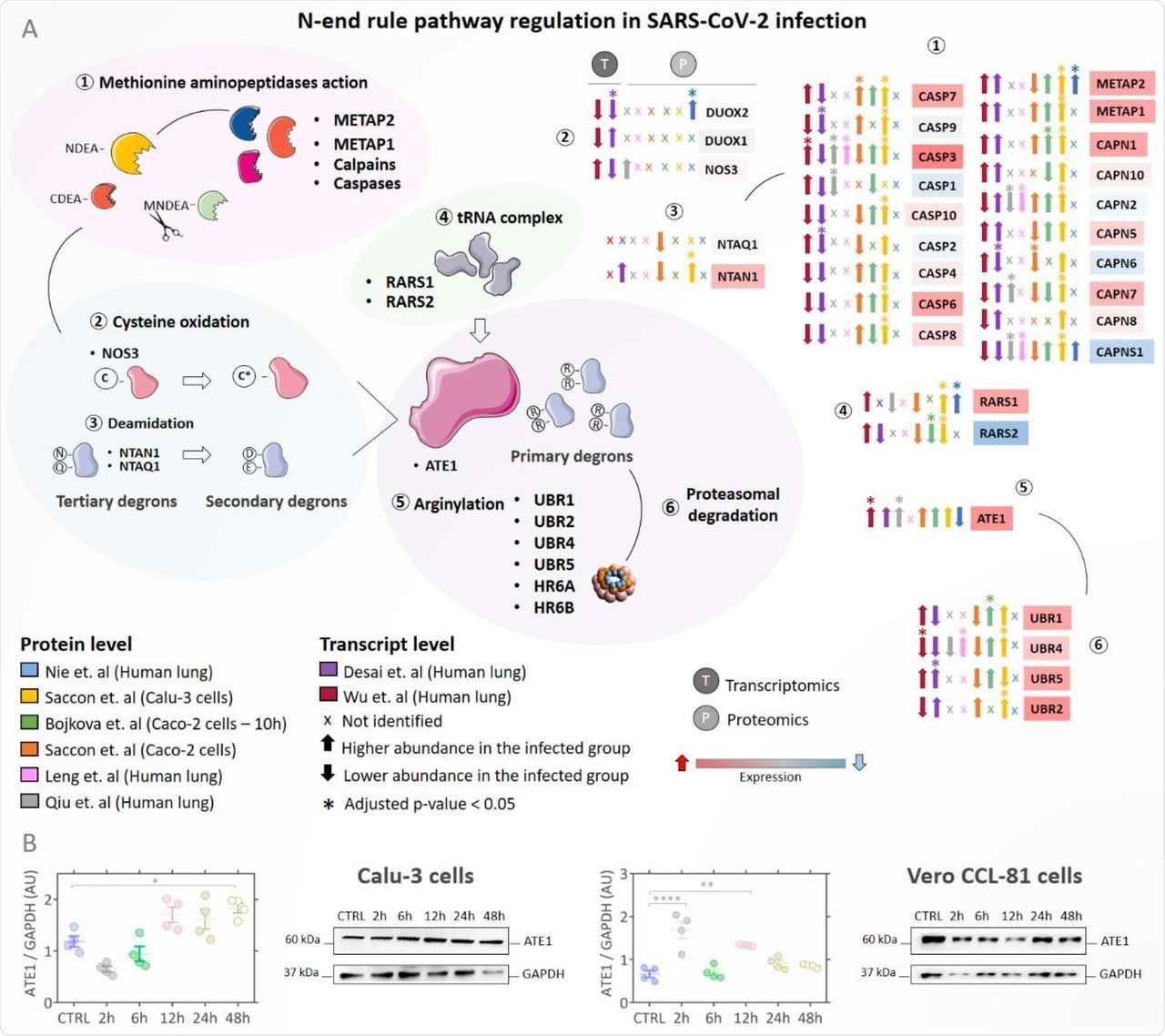 (A) Regulation of proteins involved in the N-end rule pathway during SARS-Cov-2 infection. Proteins/genes were considered differentially regulated if they had a q-value < 0.05 (Benjamini-Hochberg) and are indicated by the symbol (*). Up arrows indicate proteins/genes with the higher abundance in the infected group (INF) and down arrows indicate proteins with lower abundance in the infected group. The color of the boxes proteins/genes indicates the fold change (INF/CTRL) considering all studies evaluated; the red color indicates higher abundance in the infected group and the blue color lower abundance in the INF group. The symbol (x) indicates that a protein/gene was not identified in the dataset; (B) Western blotting analysis of ATE1 protein in Calu-3 and Vero CCL-81 cells infected with SARS-CoV-2 (classical strain) after 2h, 6h, 12h, 24h, and 48h.
(A) Regulation of proteins involved in the N-end rule pathway during SARS-Cov-2 infection. Proteins/genes were considered differentially regulated if they had a q-value < 0.05 (Benjamini-Hochberg) and are indicated by the symbol (*). Up arrows indicate proteins/genes with the higher abundance in the infected group (INF) and down arrows indicate proteins with lower abundance in the infected group. The color of the boxes proteins/genes indicates the fold change (INF/CTRL) considering all studies evaluated; the red color indicates higher abundance in the infected group and the blue color lower abundance in the INF group. The symbol (x) indicates that a protein/gene was not identified in the dataset; (B) Western blotting analysis of ATE1 protein in Calu-3 and Vero CCL-81 cells infected with SARS-CoV-2 (classical strain) after 2h, 6h, 12h, 24h, and 48h.
Further, the scientists looked for other organelles involved in arginylation during SARS-CoV-2 infection. Arginylation-related proteins were primarily located in the ER and cytoskeleton.
Since arginylation and SARS-CoV-2 infection were associated, enzyme inhibition assays were performed using the inhibitors tannic acid and merbromin (MER) in Calu-3 cells. Tannic acid and MER reduced ATE1 expression levels and inhibited arginylation. Moreover, tannic acid and MER reduced the viral load or prevented virus entry into the cell.
The scientists investigated which cell types express ATE1 by reanalyzing previously published data that used an infected group consisting of critically ill patients who were hospitalized for more than 20 days or who died due to the progression of the coronavirus disease 2019 (COVID-19) and a control group of uninfected people. This analysis found that the macrophages and epithelial cells expressed ATE1 in severe COVID-19 patients. However, the arginylation behavior in infected macrophages was different from that observed in Vero CCL-81 and Calu-3 cells.
Finally, the scientists verified whether modulation of the N-end rule pathway was recurrent in other respiratory viral infections or whether it was a specific signature of SARS-CoV-2. The N-end rule pathway was regulated in SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) infections but not in H1N1 influenza and respiratory syncytial virus (RSV) infections. Thus, there was an arginylation-dependent signature during infection with coronaviruses.
Conclusion
The current study is the first report of the role of protein arginylation and modulation of the N-end rule pathway during SARS-CoV-2 infection. The modulation of the N-end rule pathway differs between different types of infected cells, such as macrophages, Vero CCL-81, and Calu-3 cells. These findings suggest that arginylation inhibitors can reduce viral load.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Macedo-da-Silva, J., Rosa-Fernandes, L., de Moraes Gomes, V., et al. (2021). Protein arginylation is regulated during SARS-CoV-2 infection. bioRxiv. doi:10.1101/2021.11.02.466971. https://www.biorxiv.org/content/10.1101/2021.11.02.466971v1.full.
- Peer reviewed and published scientific report.
Macedo-da-Silva, Janaina, Livia Rosa-Fernandes, Vinicius de Morais Gomes, Veronica Feijoli Santiago, Deivid Martins Santos, Catarina Maria Stanischesk Molnar, Bruno Rafael Barboza, et al. 2023. “Protein Arginylation Is Regulated during SARS-CoV-2 Infection.” Viruses 15 (2): 290. https://doi.org/10.3390/v15020290. https://www.mdpi.com/1999-4915/15/2/290.