The coronavirus disease 2019 (COVID-19) pandemic has led to great research efforts in viral diagnostics. New methods have been developed to reduce diagnosis time and cost, increase sensitivity and specificity, and allow large-scale diagnostic methods to be carried out.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Several strategies were proposed that used massive parallel DNA sequencing for large-scale diagnostics. These methods used DNA molecular barcodes during sample preparation, pooling the samples for doing the assay “in one run,” and breaking down the results using bioinformatics processing.
These methods could perform diagnostics for more than a thousand samples per assay. However, they need specialized DNA sequencing machines, like the Ilumina sequencer, which need to be run by trained engineers, making it very costly.
In a study published on the medRxiv* preprint server, researchers at Paris-Saclay University in France describe a proof-of-concept of a large-scale diagnostics and tracking method for different variants of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) using DNA sequencing performed on the MinION Oxford Nanopore Technology (ONT). These sequencers are portable, low cost, and simple to use, and so can be used in low cost and remote settings.
Testing for SARS-CoV-2
To perform their SARS-CoV-2 diagnostics, the team combined a standard reverse-transcription assay with polymerase chain reaction (PCR) amplification to generate long DNA fragments. This allowed them to target short DNA fragments and, at the same time, take advantage of the capability of the sequencer to sequence long fragments. During the reverse transcription, a fragment unique to each sample is added in addition to the COVID-19 specific fragment, which allows identification of each sample.
During PCR amplification, another unique molecular fragment or barcode is added to each sample. This can increase the number of samples tested without having to linearly increase the number of primers.
In contrast to the Illumina sequencers that give sequences only at the end of the entire process, the Oxford Nanopore sequencers can provide sequences on a rolling basis. The team designed a method to collect sequences generated every few minutes, allowing for a more real-time analysis of the samples. Using a synthetic SARS-CoV-2 RNA control sample, the team validated their strategy, and they could detect down to 10 viral copies of the sample.
They further tested their method by running the sequencing for 72 hours for about four million sequence reads. From these, the authors were able to get 14,000 optimal sequences, which allowed them to detect RNA viral copies down to 100-fold dilution, which corresponds to 100 copies in the PCR assay.
Detecting variants
In addition, to the diagnostic testing, the method can also be used to detect variants in a single assay. The researchers used a combination of human RNA from cell line cultures and the synthetic RNA, including synthetic RNA corresponding to the B.1.1.7 and HF2393 SARS-CoV-2 strains.
For detecting viral variants, the sequencing focused on the SARS-CoV-2 SPIKE gene rather than the entire virus genome because most of the variations were located in the spike region. This allowed them to screen more samples, about 10 times the current sequencing coverage, making the process faster.
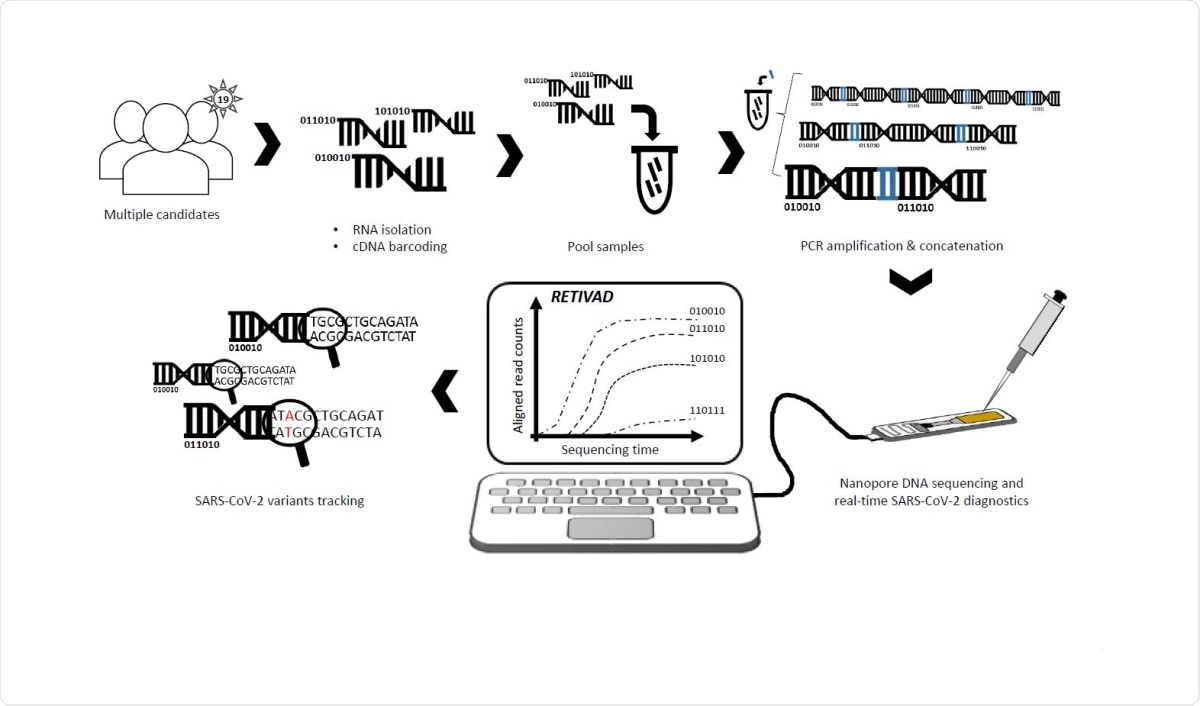
Scheme illustrating the proposed real-time SARS-CoV-2 diagnostics and variants tracking strategy. RNA samples isolated from multiple candidates are converted into complementary DNA (cDNA) such that samples associated to each candidate are labelled by molecular DNA barcodes appended to their extremities. Barcoded-cDNA samples are pooled together and PCR amplified in presence of a bridge sequence (blue) allowing to generate concatemers. Long DNA concatemers are loaded into the MinION nanopore sequencer for real-time SARS-CoV-2 diagnostics and variants tracking with the help of RETIVAD.
The negative control samples had the least number of aligned counts for the SPIKE and the E-Sarbeco (primers that define the E gene of the virus) region. Meanwhile, the samples with the highest viral RNA concentrations showed the highest number of reads aligned to these regions.
Another approach described recently for detecting variants using sequencing used 42 primer pairs to generate fragments of 400 nucleotides. The technology developed using the Nanopore sequencer uses only four primers targeting the SPIKE region, thus greatly reducing the number of reagents required.
The authors also used a computational approach they call Real-time Variants Detector (RETIVAD) to provide the diagnostic outcome in real-time. RETIVAD detected several mutations in the samples tested, including the B.1.1.7 (also known as the UK variant).
Thus, the technology described in this study is able to detect and track SARS-CoV-2 variants simultaneously in a much shorter time and uses the smaller and cheaper Oxford Nanopore sequencer, which allows the method to be used anywhere in a much affordable manner.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources