Researchers in the United States, Canada, and the UK have developed a potential new approach to predicting severe and critical outcomes among patients with coronavirus disease 2019 (COVID-19).
Brent Richards from McGill University in Montréal and colleagues say that a major clinical challenge during the COVID-19 pandemic has been the identification of individuals who are most likely to require hospitalization.
Predicting the severity of COVID-19 outcomes is complex, and the biological pathways involved are not fully understood, they say.
In a large-scale study conducted across two countries, the team has identified blood circulating proteins associated with severe COVID-19 outcomes.
The proteins identified were enriched for cytokine signaling and immune pathways, but more than half of the enriched pathways were not related to immune responses.
This suggests that many biological pathways influencing the severity of COVID-19 outcomes may act distinctly from the cytokine and chemokine proteins already known to be involved.
The researchers say the findings suggest that circulating proteins measured during the early stages of disease are reasonably accurate predictors of severe COVID-19 clinical outcomes.
“Further research is needed to understand how to incorporate protein measurement into clinical care,” they write.
A pre-print version of the research paper is available on the medRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Most biological pathways that influence COVID-19 outcomes remain unknown
A remarkable feature of COVID-19 is its highly variable clinical course, which ranges from asymptomatic to severe and fatal disease. While certain clinical and genetic risk factors account for this variation, most of the host biological pathways that influence disease outcomes remain unknown.
Therefore, the triaging of patients to identify those most likely to require hospitalization has been a major clinical challenge throughout the pandemic.
Recent reports have identified some of the biological pathways as related to immune responses, interferon pathways, and T-cell function.
However, most of these studies have focused on narrow sets of pre-selected circulating cytokines and chemokines.
Richards and colleagues say that one way to assess thousands of potential biomarkers for severe COVID-19 rapidly is to measure blood circulating proteins involved in pathways that influence the disease course.
Furthermore, circulating proteomic biomarkers have recently been shown to serve as predictors of other common diseases such as cardiovascular disease.
These biomarkers are also relevant in drug discovery since they are more accessible to pharmacological manipulation than intracellular proteins.
Understanding the circulating proteins associated with adverse COVID-19 outcomes may help address some of the significant challenges faced in the current pandemic.
What did the researchers do?
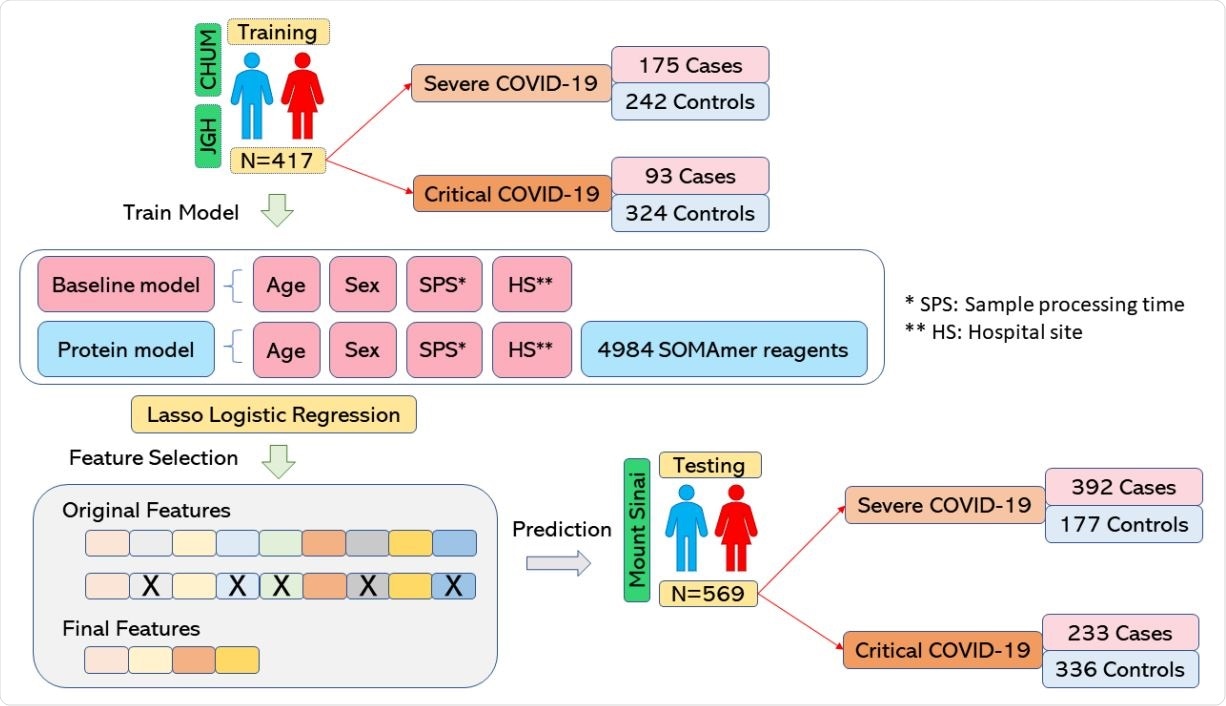
Richards and colleagues conducted a large-scale study assessing the association between thousands of circulating proteins and severe and critical COVID-19 outcomes. They used machine learning to develop a predictive model of COVID-19 severity using the circulating blood proteins as predictors.
Almost 5,000 nucleic acid aptamers targeted 4,701 unique circulating proteins in two independent cohorts from two countries, totaling 986 individuals.
The team trained prediction models, including protein abundances and clinical risk factors, to predict adverse outcomes in 417 individuals and then tested the performance of these models in a separate external cohort of 569 individuals.
Point-of-Care (POC) device detects SARS-CoV-2 in patient samples (A) Signal acquisition module of the POC device with main elements labeled. (B) Photograph of the gFET Cartridge Unit and the Signal acquisition modules connected together to form the entire POC. A reference dimension bar is reported below. (C) Schematic representation of gFET modified with ACE2-Fc tested on viral samples from patients. (D) Bar graph reporting ACE2-Fc_gFET signal before (black) and after the addition of swab specimens (red) from patient 1, 2 and 3. **P<.01, ***P<.001; (E) RT-qPCR results for the detection of the SARS-CoV-2 specific genes (E gene; RdRP gene; S gene and ORF1ab gene) of the three patient samples. Ct value in clinical samples were evaluated. Ct average value of internal control are indicated for each sample. The type of virus variant (as detected by targeted RT-PCR) is also reported.
A subset of proteins were strong predictors of COVID-19 severity
The researchers identified a subset of proteins that were strong predictors of COVID-19 severity. The proteomic model was able to predict severe COVID-19 – defined as requiring the use of oxygen – with a positive predictive value of 89%.
The proteins identified were strongly enriched for cytokine signaling and immune pathways, but they also highlighted pathways that were not immune-related.
Interestingly, only 5 of 14 proteins that were selected in the final model of both severe and critical outcomes were cytokines or chemokines.
The cytokine IFNA7 and the chemokines CXCL13, CXCL10, CCL7, and CCL8 were selected for predicting severe outcomes, while CXCL13, CXCL10, and CCL7 were also selected for predicting critical outcomes.
Other proteins selected were specific to non-immune response pathways such as protein phosphorylation and glycosaminoglycan binding, pointing to biological pathways that act distinctly from known cytokine and chemokine proteins.
Could this proteomic approach assist in triaging patients for hospital admission?
The researchers say the findings provide insights into the biological pathways that influence COVID-19 outcomes and the ability of proteomics to predict these outcomes.
Circulating protein levels are strongly associated with these outcomes and are able to predict the need for oxygen supplementation or death with reasonable accuracy, says the team.
“Further research is needed to assess whether this proteomic approach can be applied in a clinical setting to assist in triaging patients for admission to hospital,” concludes Richards and colleagues.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Richards B, et al. Circulating proteins to predict adverse COVID-19 outcomes. medRxiv, 2021. doi: https://doi.org/10.1101/2021.10.04.21264015, https://www.medrxiv.org/content/10.1101/2021.10.04.21264015v1
- Peer reviewed and published scientific report.
Su, Chen-Yang, Sirui Zhou, Edgar Gonzalez-Kozlova, Guillaume Butler-Laporte, Elsa Brunet-Ratnasingham, Tomoko Nakanishi, Wonseok Jeon, et al. 2023. “Circulating Proteins to Predict COVID-19 Severity.” Scientific Reports 13 (1): 6236. https://doi.org/10.1038/s41598-023-31850-y. https://www.nature.com/articles/s41598-023-31850-y.