The worldwide spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in an economic and global health crisis. For a host to be protected from SARS-CoV-2 infection or the development of severe disease, coordinated activation of different immune system components is needed. A successful immune response, therefore, leads to the production of antigen-binding and neutralizing antibodies, as well as antiviral T-cells.
 Study: Rapid measurement of SARS-CoV-2 spike T cells in whole blood from vaccinated and naturally infected individuals. Image Credit: CI Photos / Shutterstock.com
Study: Rapid measurement of SARS-CoV-2 spike T cells in whole blood from vaccinated and naturally infected individuals. Image Credit: CI Photos / Shutterstock.com
SARS-CoV-2 spike protein-specific antibodies and T-cells are induced by the recently developed SARS-CoV-2 vaccines, several of which provide protection in about 90% of fully vaccinated individuals. However, it is not yet fully known what level of antibodies and/or T-cells is necessary to provide effective protection against SARS-CoV-2 infection.
Recently, a group of researchers applied cellular methods to measure SARS-CoV-2 T-cell responses in individuals vaccinated with the BNT162b2 vaccine or in those who developed immunity from a prior natural infection. In their study, published in the Journal of Clinical Investigation, the authors show that through the addition of spike peptide pools to whole blood, the detection and quantification of spike-specific T-cells can be easily achieved.
Rapid quantification of SARS-CoV-2 spike-specific T-cells
Through the use of different methods of antigen-specific T-cell analysis in fresh blood as well as cryopreserved peripheral blood mononuclear cells (PBMCs), the authors characterized the initial kinetics of spike-specific T-cells gained from two doses of the BNT162b2 messenger ribonucleic acid (mRNA) vaccine over a period of 51 days.
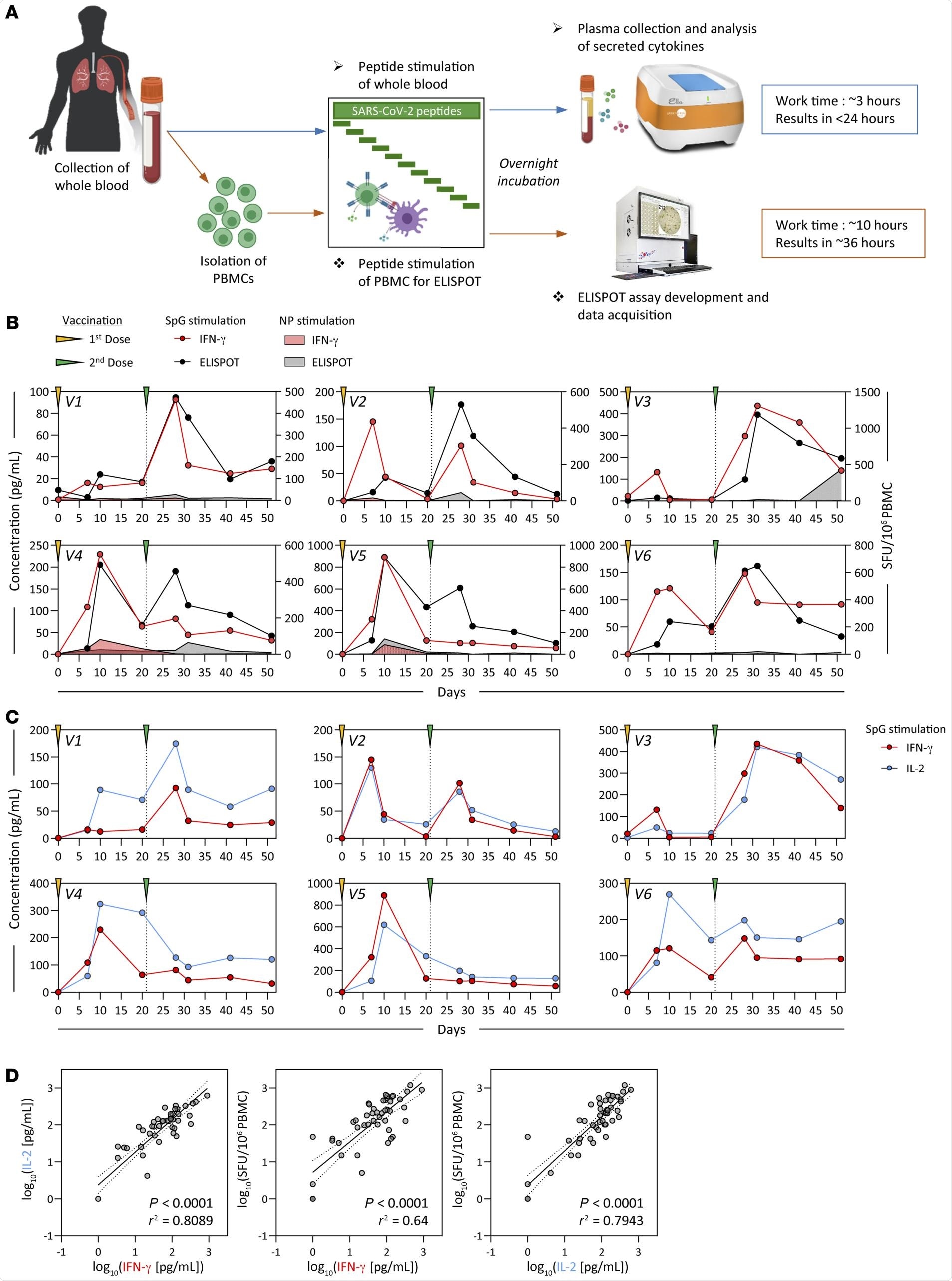 Detection of SARS-CoV-2 spike–specific T cells by peptide stimulation of whole peripheral blood from vaccinated individuals. (A) Schematic representation of the workflow for the direct peptide stimulation of whole peripheral blood and the subsequent detection of cytokine secretion compared with a standard IFN-γ ELISPOT assay. (B) Six healthy individuals were vaccinated with 2 doses of BNT162b2 according to the recommended schedule (21 days apart), and whole-blood samples were longitudinally analyzed 7, 10, 20, and 30 days after each dose. The collected whole blood was either directly stimulated for 16 hours with peptide pools specific for the spike protein (red or black line) or NP (red or black shaded area), or immediately processed with Ficoll density gradient centrifugation to isolate PBMCs. A standard IFN-γ ELISPOT assay using the SpG- or NP-specific peptide pools was then set up using the freshly isolated PBMCs. The quantity of secreted IFN-γ in stimulated whole blood (red line) was compared with the frequency of peptide-reactive PBMCs quantified by IFN-γ ELISPOT (black line). (C) The levels of secreted IL-2 (blue line) in whole blood stimulated with the SpG peptide pool were compared with the amount of IFN-γ detected. (D) Linear regression analysis of the concentrations of IFN-γ and IL-2 in SpG-specific peptide pool–stimulated whole blood and the corresponding frequency of spike-specific PBMCs (n = 6; 48 samples). Dotted lines denote the 95% CI.
Detection of SARS-CoV-2 spike–specific T cells by peptide stimulation of whole peripheral blood from vaccinated individuals. (A) Schematic representation of the workflow for the direct peptide stimulation of whole peripheral blood and the subsequent detection of cytokine secretion compared with a standard IFN-γ ELISPOT assay. (B) Six healthy individuals were vaccinated with 2 doses of BNT162b2 according to the recommended schedule (21 days apart), and whole-blood samples were longitudinally analyzed 7, 10, 20, and 30 days after each dose. The collected whole blood was either directly stimulated for 16 hours with peptide pools specific for the spike protein (red or black line) or NP (red or black shaded area), or immediately processed with Ficoll density gradient centrifugation to isolate PBMCs. A standard IFN-γ ELISPOT assay using the SpG- or NP-specific peptide pools was then set up using the freshly isolated PBMCs. The quantity of secreted IFN-γ in stimulated whole blood (red line) was compared with the frequency of peptide-reactive PBMCs quantified by IFN-γ ELISPOT (black line). (C) The levels of secreted IL-2 (blue line) in whole blood stimulated with the SpG peptide pool were compared with the amount of IFN-γ detected. (D) Linear regression analysis of the concentrations of IFN-γ and IL-2 in SpG-specific peptide pool–stimulated whole blood and the corresponding frequency of spike-specific PBMCs (n = 6; 48 samples). Dotted lines denote the 95% CI.
Two different assays revealed a principal spike-specific response in all individuals and defined a matching profile of spike-specific T-cell responses following the first and second vaccinations. Following the second vaccination, the number of interferon-gamma (IFN-y) spots detected were similar to the numbers observed in the clinical trials that included individuals who had received the BNT162b2 vaccine.
Furthermore, the researchers found a similar preparation consistency of the trimerized secreted version of the spike receptor-binding domain to arise.
T-cells specific for the SpG peptide pool
Information on the immunogenicity of the different regions of the spike protein and the total spike-specific T-cell responses were obtained through the use of a whole blood cytokine release assay (CRA).
Comparatively, the assessment of the response in large numbers of individuals required a more streamlined approach. Thus, the authors examined the relationship between the total spike-specific T-cell response and the response to their selected SpG peptide pool.
The authors observed a strong positive linear relationship demonstrating that the T-cell response against the SpG peptide pool highly represented the total spike T-cell response. This observation was based on the correlation of results from three different assays, in which both the SpG peptide pool-specific T-cell responses and the total spike protein were determined in the same sample through stimulation with the corresponding peptide pools..
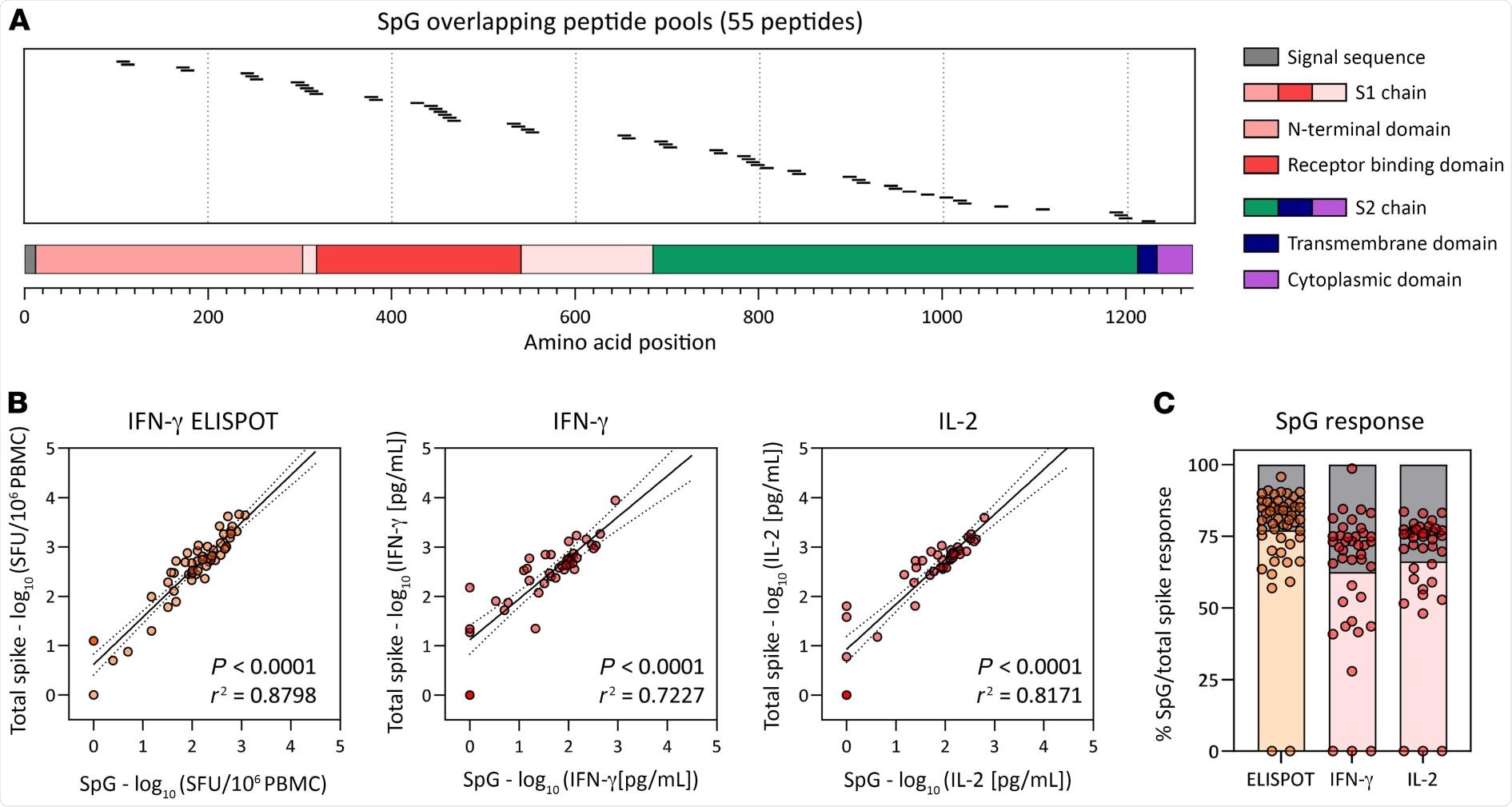 Frequency of the SpG peptide pool and total spike protein–specific T cells. (A) Schematic representation of the individual 15 mer overlapping peptides contained in the SpG peptide pool. (B) Linear regression analysis of the T cell response against the SpG peptide pool and the total spike protein (pools 1–7) as evaluated by ELISPOT (left) or by the quantification of IFN-γ (middle) or IL-2 (right) in peptide-stimulated whole blood (n = 6; 42 samples). (C) The SpG peptide pool–specific T cell response quantified by each assay is expressed as a fraction of the total spike protein T cell response observed (n = 6; 42 samples). Bars indicate the mean.
Frequency of the SpG peptide pool and total spike protein–specific T cells. (A) Schematic representation of the individual 15 mer overlapping peptides contained in the SpG peptide pool. (B) Linear regression analysis of the T cell response against the SpG peptide pool and the total spike protein (pools 1–7) as evaluated by ELISPOT (left) or by the quantification of IFN-γ (middle) or IL-2 (right) in peptide-stimulated whole blood (n = 6; 42 samples). (C) The SpG peptide pool–specific T cell response quantified by each assay is expressed as a fraction of the total spike protein T cell response observed (n = 6; 42 samples). Bars indicate the mean.
Implications
In the current study, the authors show that a wide and dynamic range of functionally heterogenous spike-specific T-cell responses that are induced through vaccination or infection can be rapidly detected through the measurement of cytokine production in whole blood after peptide-specific activation via an assay.
Despite T-cells being unable to prevent infection without the presence of antibodies, they play a key role in protecting people from developing severe disease. This has been observed in individuals who acquire SARS-CoV-2 via natural infection, as well as those who have been vaccinated.
Journal reference:
- Tan, A. T., Lim, J. M. E., Le Bert, N., et al (2021). Rapid measurement of SARS-CoV-2 spike T cells in whole blood from vaccinated and naturally infected individuals. The Journal of Clinical Investigation. doi:10.1172/JCI152379.