A recent study reveals that the genetic variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detected in the initial phase of coronavirus disease 2019 (COVID-19) pandemic differ from the variants detected in a massive second wave of infection in Houston, Texas. The study is currently available on the medRxiv* preprint server.
According to the World Health Organization (WHO), the rapid spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in over 32 million confirmed COVID-19 cases, including 979,000 deaths worldwide, as of 25 September 2020. In the United States, the regions worst hit by the COVID-19 pandemic include Seattle and New York City.
In the Houston metropolitan area, which is the 4th largest and ethnically diverse city in the United States, the first confirmed case of COVID-19 was registered on 5 March 2020, and the community transmission of SARS-CoV-2 started a week later.
In Houston, a continuous effort has been made to identify positive COVID-19 cases and evaluate genomic variants of SARS-CoV-2 since the initial phase of the pandemic. The effort has been significantly expanded with the emergence of a second wave of SARS-CoV-2 infection in late May 2020.
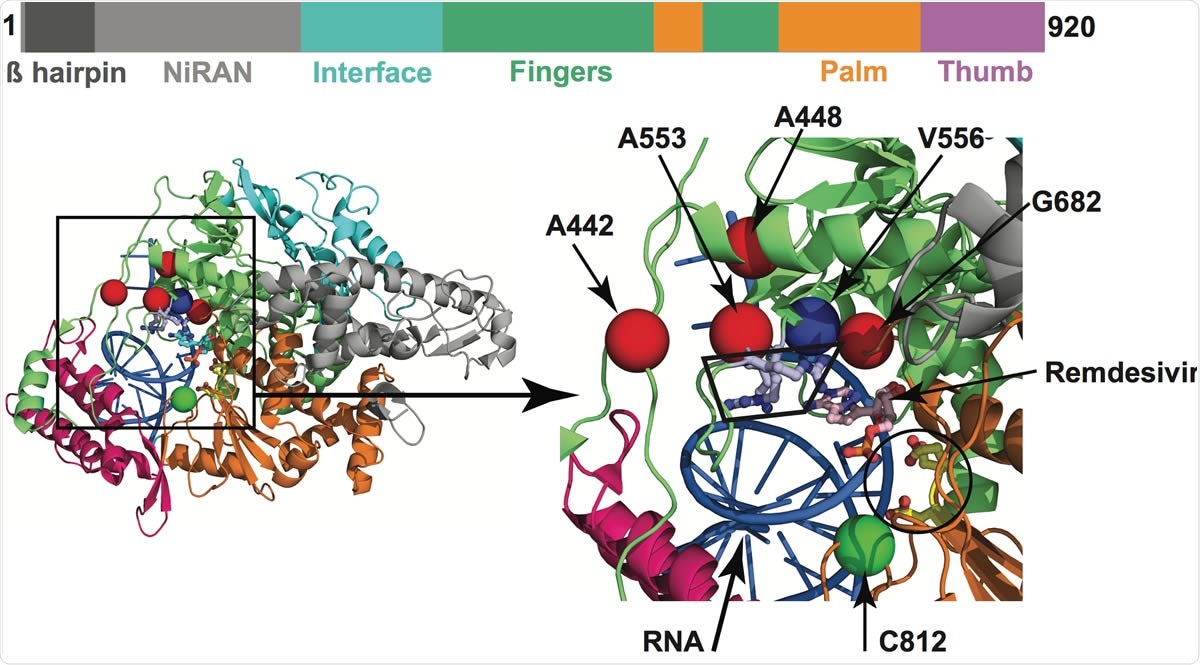
Amino acid changes identified in Nsp12 (RdRp) in this study that may influence interaction with remdesivir. The schematic at the top shows the domain architecture of Nsp12. (Left) Ribbon representation of the crystal structure of Nsp12-remdesivir monophosphate-RNA complex (PDB code: 7BV2). The structure in the right panel shows a magnified view of the boxed area in the left panel. The Nsp12 domains are colored as in the schematic at the top. The catalytic site in Nsp12 is marked by a black circle in the right panel. The side chains of amino acids comprising the catalytic site of RdRp (Ser758, Asp759, and Asp760) are shown as balls and stick and colored yellow. The nucleotide binding site is boxed in the right panel. The side chains of amino acids participating in nucleotide binding (Lys544, Arg552, and Arg554) are shown as balls and sticks and colored light blue. Remdesivir molecule incorporated into the nascent RNA is shown as balls and sticks and colored light pink. The RNA is shown as a blue cartoon and bases are shown as sticks. The positions of Ca atoms of amino acids identified in acids that are shown as red spheres are located above the nucleotide binding site, whereas Cys812 located at the catalytic site is shown as a green sphere. The side chain of active site residue Ser758 is shown as ball and sticks and colored yellow. The location of Ca atoms of remdesivir resistance conferring amino acid Val556 is shown as blue sphere and labeled.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Current study objective
The current study by researchers from the Houston Methodist Hospital, University of Chicago, and the University of Texas at Austin was designed to evaluate the viral introduction events in the Houston metropolitan area and identify the genomic variants of SARS-CoV-2 that have infected people during the initial wave and a second prominent wave of SARS-CoV-2 infection.
Study design
The COVID-19 trajectory graph demonstrated that the number of SARS-CoV-2 infected people peaked around mid-April, followed by a reduction in case numbers until 11 May. However, there was a sharp increase in case numbers beginning on 12 June. Considering this trajectory, the scientists defined the 1st wave of infection as 5 March – 11 May 2020, and the 2nd wave as 12 May – 7 July.
In the early phase of the 1st wave, they tested 3,080 patient samples and found 406 SARS-CoV-2 positive cases. These cases represent 40% of all confirmed cases in the Houston metropolitan area. During the entire study period, they identified 9,121 SARS-CoV-2 positive cases out of 55,800 suspected cases. These cases represent 17% of all confirmed cases in the metropolitan area. According to the scientists, the viral strains they identified represent all COVID-19 cases during the 1st and 2nd waves of infection.
Important observations
By analyzing the characteristics of patients, the scientist observed that the people affected in the second wave were much younger, had lower numbers of comorbidities, lived in low-income families, and was mostly Hispanic/Latino.
Regarding genetic variants of SARS-CoV-2, the scientists identified a diverse-range of viral genomes in the 1st wave, and the majority of these strains belonged to the clades G, GH, GR, and S. These are the major genetic clades causing COVID-19 globally.
About 82% and 99.9% of the viral strains in the 1st and 2nd waves had D614G mutation in the spike protein, respectively. The D614G mutation has been shown to increase the transmission efficiency and infectivity of SARS-CoV-2.
The presence of different variants of SARS-CoV-2 in the early phase of 1st wave indicates several independent viral entry events in Houston instead of the introduction and transmission of a single variant.
Using machine learning models, they observed that the acquisition of D614G mutation was not correlated with the disease outcomes. However, people infected with the mutated variant of SARS-CoV-2 showed significantly higher viral load in the nasopharynx.
Analysis of the genetic sequence coding for the spike protein identified 470 single nucleotide polymorphisms, of which 285 caused amino acid changes. These amino acid substitutions were identified in each spike protein domain and subunit. The most frequently occurring substitution was D614G.
Furthermore, the scientists generated a spike protein construct carrying D614G mutation and observed that the mutated spike protein has higher thermostability and increased expression level. Pseudotyped viruses with the D614G variant of the spike protein showed significantly higher infectivity for host cells in vitro.
Additionally, they created 13 receptor-binding domain (RBD) variants carrying D614G mutation along with an additional single amino acid substitution that had been identified in the genome analysis data.
They observed that the majority of these variants have a high affinity for the angiotensin-converting enzyme (ACE) receptor.
Interestingly, they observed that of these variants, three (F338L, S373P, and R408T) had significantly reduced affinity for a monoclonal neutralizing antibody, namely CR3022, which is known to disrupt the spike protein homotrimerization interface.
Of these variants, S373P is located one amino acid away from the CR3022-specific epitope. These observations indicate that SARS-CoV-2 acquiring mutations in the RBD can potentially escape the antibody-mediated host immune responses.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources