Ampli1™ LowPass Kit applications include the characterization and investigation of genomic heterogeneity and evolution in tumor cell populations such as individual circulating tumor cells (CTCs).
Ampli1™ LowPass Kit is designed to generate multiplexed, sequencing-ready libraries in a streamlined single-day protocol, to detect chromosomal aneuploidies and copy number aberrations (CNAs) by low-pass whole genome sequencing.
Ampli1™ LowPass Kit is compatible with Ion Torrent systems (Ion PGM™ and Ion S5™) and NOW also compatible with Illumina® systems (MiSeq®, HiSeq® 1000/1500, 2000/2500, 3000/4000 and NovaSeq™).
The Ampli1™ LowPass Kit works exclusively with Ampli1™ WGA products. The complete workflow takes advantage of the isolation of 100% pure cells with the DEPArray™ system and balanced whole genome amplification using the Ampli1™ WGA kit.

Performance of this method has been extensively validated by analyzing the Copy Number profiles of single CTCs and WBCs from cancer patients (see figure below for representative data).


Low-pass Whole Genome Profile (chr 1-22) of single CTC (upper panel) and WBC (lower panel) isolated with DEPArray™ Technology from a patient with Lung Adenocarcinoma. Ploidy values are indicated in the y-axis; in the x-axis the alternation of different chromosomes are plotted with different colors. CNAs in the CTC are indicated in red and blue.
Ampli1™ LowPass Kit for Ion Torrent - Overview
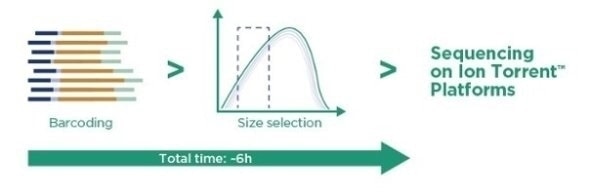
- Two-step procedure that avoids the laborious fragmentation step and preparation of several enzyme reactions.
- Up to 96 barcoded libraries suitable for both Ion Torrent NGS platforms: Ion PGM™ and Ion S5™.
- Accurate characterization of CNA profiles of multiple single tumor cells per single run.
- Fast results.
Ampli1™ LowPass Kit for Illumina - Overview
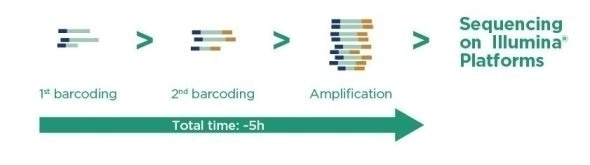
- Three-step procedure based on a novel, fragmentation-free and size selection-free method.
- Single-tube prep method.
- Up to 96 barcoded libraries suitable for Illumina MiSeq®, HiSeq® 1000/1500, 2000/2500, 3000/4000 and NovaSeq™ sequencing instruments.
- Suitable for high-throughput liquid handling automation.
References
- Buson G et al, AACR 2016, “Scalable, rapid and affordable low-pass whole genome sequencing method for single-cell copy-number profiling on LM-PCR based WGA products”
- Ferrarini A et al, AACR 2016, “Precise copy-number profiling of single cells isolated from FFPE tissues by low-pass whole-genome sequencing”