Most previously known anti-SARS-CoV-2 antibodies block receptor-binding-domain (RBD)-binding to the host angiotensin-converting enzyme 2 (ACE2)-receptor and prevent the endocytosis step of SARS-CoV-2 entry. However, SP1-77 is unique because it binds the RBD of the SARS-CoV-2 spike (S) protein via novel complementarity-determining-region-3(CDR3) sequences to block viral and host cell membrane fusion.
The number of primary B cells in the immune system determines the primary B-cell repertoire (BCR) in humans and mice. A mouse has 1000-fold fewer B cells than a human; therefore, they express a tiny fraction of the CDR3 diversity compared to humans. Preliminary experimental data indicated rearrangement of a single human variable region of heavy (VH) regions of immunoglobulin (Ig) and kappa light-chain variable region (Vκ) of Ig could generate BCR repertoires capable of responding to diverse antigens due to immense CDR3 diversification.
About the study
In the present study, researchers used a humanized mouse model that generated more human-like CDR3 diversity to yield therapeutic mAbs that could neutralize several SARS-CoV-2 VOCs. This mouse model rearranged a single human VH gene segment (VH1-2) and Vκ gene segment (Vκ1-33).
They vaccinated these mice with recombinant stabilized soluble SARS-CoV-2 Wuhan-1 S trimer, or an RBD monomer fused to a nanobody "VHH7-RBD" twice, four weeks apart. Serum IgG titers at two and six weeks post-vaccination revealed all mice developed strong antibody responses to the SARS-CoV-2 S or RBD immunogens. After three weeks of the second dose, the team single-cell sorted fluorescence-activated 96 antigen-specific IgG+ B cells from each mouse. Next, they sequenced their Ig heavy chain (IgH) and Ig light chain (IgL) variable region exons.
Study findings
Since the humanized mice had human VH1-2 and Vκ1-33, they contributed to the BCRs expressed by 100% and 11% of splenic B cells, respectively. Cytofluorimetry showed that splenic B and T cell populations in this mouse model were similar in numbers and characteristics to the wild-type mice. Additionally, they had a diverse VH1-2- and Vκ1-33-based CDR3 repertoire.
The study experiments yielded nine S-specific B cell lineages, each expressing BCRs with unique CDR3s. In addition, an enzyme-linked immunosorbent assay (ELISA) identified six mAbs bound to RBD, with one termed SP1-77 potently neutralizing all currently known SARS-CoV-2 variants. It also robustly neutralized the recently emerging Omicron sub-variants, BA.1, BA.2, BA.3, BA.4/BA.5, and BA.2.12.1.
In a plaque reduction neutralization test (PRNT) on the live SARS-CoV-2 virus, SP1-77 had the half-maximal inhibitory concentration (IC50) values ranging from 0.8 to 9.6 ng/ml. Surface plasmon resonance (SPR) confirmed SP1-77 bound the back and right faces outside the receptor-binding motif (RBM) and did not compete with ACE2 for binding to the RBD or S trimer.
The cryo-electron microscopic (cryo-EM) structure of the full-length G614 S trimer in complex with the SP1-77 fragment antigen-binding region (Fab) validated the SPR findings. SP1-77 directly contacted the RBD, but also touched the N-terminal domain (NTD) from a neighboring protomer. The SP1-77-RBD interaction buried a 939 square angstroms (Å2) surface area on the antibody and a 936 Å2 area from the S protein. Furthermore, the IgH and IgL of SP1-77 contributed to 83% and 17% of the binding surface area, respectively. The Glycine339Asp and Asn440Lys mutations were at the edge of the SP1-77 epitope with their side chains pointing away from the binding interface, explaining why Omicron variants were susceptible to SP1-77 neutralization.
The researchers used a lattice light-sheet microscopy (LLSM) approach to demonstrate the SP1-77 neutralization mechanism. While pre-incubation of viruses with SP1-77 had a minimal impact on viral endocytosis, it dramatically inhibited fluorescence spreading within the endosome, indicative of viral-endosomal membrane fusion block. Together, these findings suggested that SP1-77 neutralized SARS-CoV-2 by blocking membrane fusion.
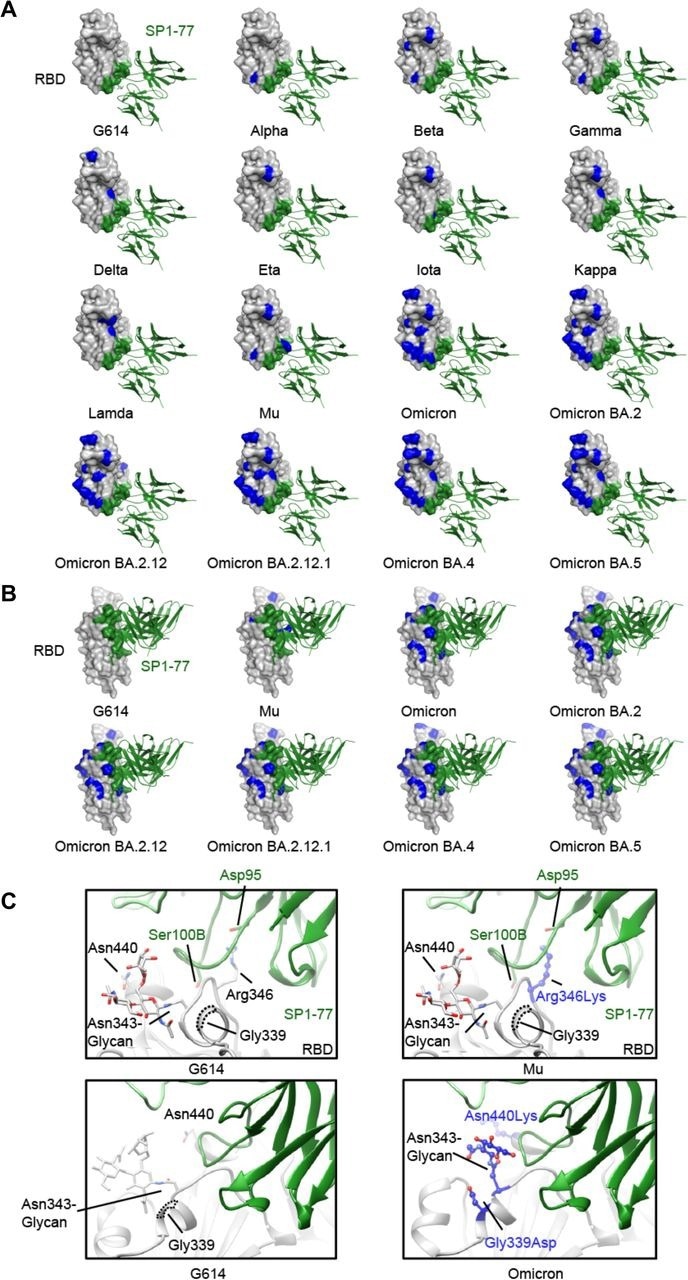
Modeled SP1-77 binding site on various SARS-CoV-2 variants. (A) The potential footprint of SP1-77 on the modeled RBDs from different SARS-CoV-2 variants in a top view. The RBD is shown in surface representation in gray with the SP1-77 footprint highlighted in green and the mutations in each variant in blue. The Fv region of SP1-77 is shown in ribbon diagram in green. Most mutations on spike variants are not located at the SP1-77 footprint. (B) Side view of a selected panel from A. (C) Structural comparison of the SP1-77 binding interface among the RBDs of wildtype G614, Mu and Omicron variants. The conservative nutation R343K in Mu preserves the salt bridge between the residue 343 in the RBD and the SP1-77 D99. The mutations in Omicron variant reconfigures the local conformation near the N343-glycan, which is on the edge of SP1-77 footprint. The RBD is colored in gray, the SP1-77 heavy chain in green and the light chain in light green. Mutations in the Mu and Omicron variants are shown in stick and ball model in blue.
Conclusions
The study provides the first insight into how a non-ACE2 blocking antibody can potently neutralize multiple SARS-CoV-2 variants. The unique binding characteristics and novel neutralization mechanism of the SP1-77, coupled with its broad and potent neutralization of SARS-CoV-2 variants, suggested that it could be therapeutic against current and yet to emerge VOCs. Also, SP1-77 might be used in cocktail mAb therapies, e.g., with LY-CoV1404, that neutralize multiple SARS-CoV-2 VOCs through an ACE2-blocking mechanism. Additionally, its novel neutralization mechanism could inspire the design of new vaccines targeting Omicron BA.2 and other VOCs.
Finally, the study highlighted the success of the VH1-2/Vκ1-33-rearranging mouse model as a prototype for testing humanized antibodies and vaccine strategies against other diverse pathogens, such as human immunodeficiency virus-1 (HIV-1) and influenza.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Humanized antibody potently neutralizes all SARS-CoV-2 variants by a novel mechanism, Sai Luo, Jun Zhang, Alex J.B. Kreutzberger, Amanda Eaton, Robert J. Edwards, Changbin Jing, Hai-Qiang Dai, Gregory D. Sempowski, Kenneth Cronin, Robert Parks, Adam Yongxin Ye, Katayoun Mansouri, Maggie Barr, Novalia Pishesha, Aimee Chapdelaine Williams, Lucas Vieira Francisco, Anand Saminathan, Hanqin Peng, Himanshu Batra, Lorenza Bellusci, Surender Khurana, S. Munir Alam, David C. Montefiori, Kevin O. Saunders, Ming Tian, Hidde Ploegh, Tom Kirchhausen, Bing Chen, Barton F. Haynes, Frederick W. Alt, bioRxiv pre-print 2022, DOI: https://doi.org/10.1101/2022.06.26.497634, https://www.biorxiv.org/content/10.1101/2022.06.26.497634v1
- Peer reviewed and published scientific report.
Luo, Sai, Jun Zhang, Alex J.B. Kreutzberger, Amanda Eaton, Robert J. Edwards, Changbin Jing, Hai-Qiang Dai, et al. 2022. “An Antibody from Single Human v H -Rearranging Mouse Neutralizes All SARS-CoV-2 Variants through BA.5 by Inhibiting Membrane Fusion.” Science Immunology, August. https://doi.org/10.1126/sciimmunol.add5446. https://www.science.org/doi/10.1126/sciimmunol.add5446.