Overview of Hi-Gx360
Hi-Gx360 enables companies to improve their genomics and bioinformatics workflow, ensuring a smooth trip from sample to insights.
Hi-Gx360® sequencing and bioinformatics services
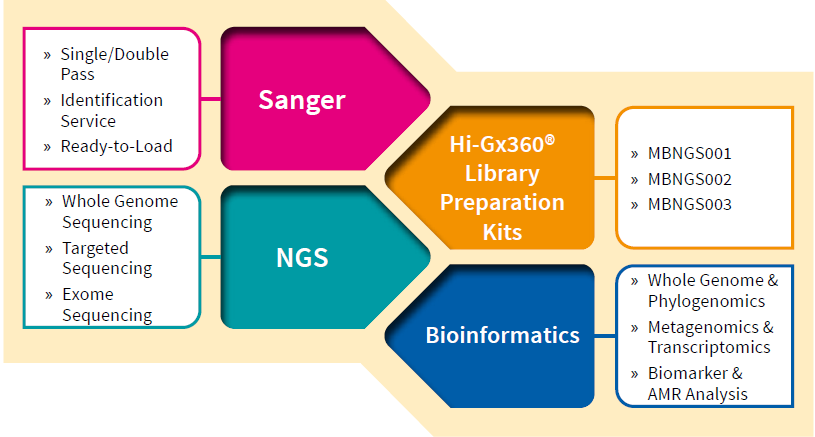
Image Credit: HiMedia Laboratories Pvt. Ltd.
HiMedia stands for:
- High-quality data production for many sequencing applications
- No hidden fees
- Standard vector primers or universal gene primers already available with us can be offered (only for sequencing services) at no additional expense
- No additional price for difficult-to-sequence templates
- Help debug sequencing tasks at no additional cost
- Completely customized solutions that meet the project requirements (based on prior discussion)
- Analytic pipelines are functionally evaluated, optimized, and verified utilizing open source technologies
- Experts in bioinformatics and sequencing provide direct support
- Shared knowledge base and resources for user empowerment
State-of-art Hi-Gx360® lab


Image Credit: HiMedia Laboratories Pvt. Ltd.
Sanger sequencing applications
- A very accurate 'chain termination-based' 'first generation' DNA sequencing technique
- The gold standard for benchmarking all other sequencing methods, including NGS
- Confirm sequence variants or fill ‘gaps’ in genomic areas identified by NGS
- Predesigned identification service under Isolate-to-Identity, encompassing all organisms
- Detailed reports always contain .ab1 files and other relevant information
- Standard vector primers and universal gene markers included at no cost (only for sequencing services)
- Gene sequence-based phylogenetic analysis service
- Stuck with NCBI GenBank submissions, connect with HiMedia
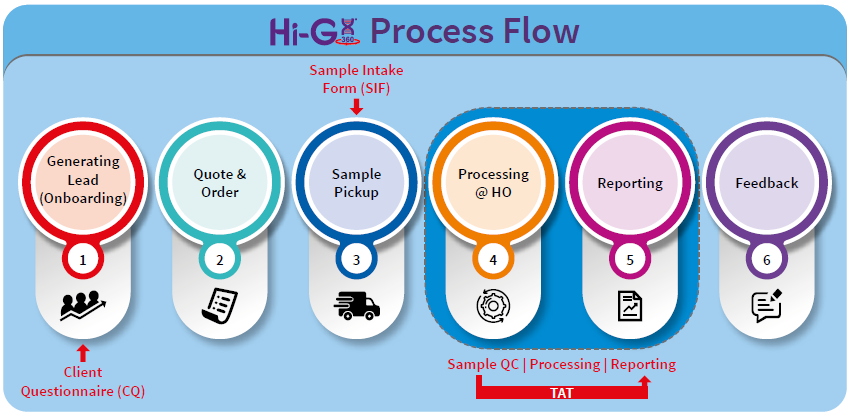
Image Credit: HiMedia Laboratories Pvt. Ltd.
Sanger sequencing services
Source: HiMedia Laboratories Pvt. Ltd.
| Description |
|
Service Codes |
| Single pass sequencing ~700 bp |
purified PCR/Plasmid |
MBS101 |
| unpurified PCR/Plasmid |
MBS102 |
| Single-strand F & R primer sequencing ~1200 bp |
purified PCR/Plasmid |
MBS103 |
| unpurified PCR/Plasmid |
MBS104 |
| Single strand F&R primer sequencing & BLAST analysis ~1200 bp |
unpurified PCR/Plasmid |
MBS104A |
| Double pass both strands 1.5-2 kb |
purified PCR/Plasmid |
MBS105 |
| unpurified PCR/Plasmid |
MBS106 |
| 96-well plate single pass sequencing ~700 bp |
purified PCR/Plasmid |
MBS107 |
| unpurified PCR/Plasmid |
MBS108 |
| Primer walking (upto 2 kb) |
|
MBS121 |
| Primer walking (upto 5 kb) with DNA extraction |
|
MBS121A |
| Ready-to-Run 96-well plate single pass sequencing (~700 bp) |
|
MBS122 |
| gDNA-to-Sequence 96-well plate single pass sequencing (~700 bp) |
|
MBS123 |
| gDNA-to-Sequence 96-well plate F & R primer sequencing (~1200 bp) |
|
MBS124 |
Supplemental services
Only available in conjunction with other service codes, not independently.
Source: HiMedia Laboratories Pvt. Ltd.
| Description |
Service Codes |
| Agarose Gel Extraction |
MBS301 |
| Plasmid DNA Isolation |
MBS302 |
| Purification of PCR Products |
MBS303 |
| Agarose Gel-based QC |
MBS305 |
Identification of organisms (Isolate-to-Identity)
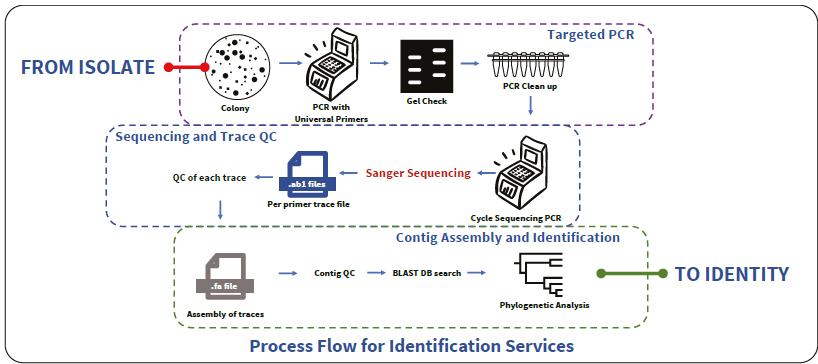
Image Credit: HiMedia Laboratories Pvt. Ltd.
Identification services
Source: HiMedia Laboratories Pvt. Ltd.
| Description |
|
Service Codes |
| Bacterial/Archaeal 16S rRNA-based Identification |
From Genomic DNA |
MBS109 |
| From Pure Isolate |
MBS110 |
16s rRNA-based partial Identification
(gDNA to BLAST, ~700 bp single pass) |
Does not include phylogenetic analysis (MBS020). |
MBS109A |
16s rRNA-based partial Identification
(gDNA to BLAST, ~1200 bp single pass) |
Does not include phylogenetic analysis (MBS020). |
MBS109B |
| Fungal ITS-based Identification |
From Genomic DNA |
MBS111 |
| From Pure Isolates |
MBS112 |
| Fungal 18S rRNA-based Identification |
From Genomic DNA |
MBS113 |
| From Pure Isolates |
MBS114 |
| Algal 18S rRNA-based Identification |
From Genomic DNA |
MBS115 |
| From Pure Isolates |
MBS116 |
| Algal 23S rRNA-based Identification |
From Genomic DNA |
MBS117 |
| From Pure Isolates |
MBS118 |
NABL accredited services
Source: HiMedia Laboratories Pvt. Ltd.
| Description |
Service Codes |
| Hi-Gx360® Microbial Identification by 16S rRNA gene sequencing from genomic DNA |
MBS409 |
| Hi-Gx360® Microbial Identification by 16S rRNA gene sequencing from pure isolate |
MBS410 |
| Hi-Gx360® Fungal Identification by ITS sequencing from genomic DNA |
MBS411 |
| Hi-Gx360® Fungal Identification by ITS sequencing from Pure Isolates |
MBS412 |
| Hi-Gx360® Fungal Identification by 18S rRNA gene sequencing from genomic DNA |
MBS413 |
| Hi-Gx360® Fungal Identification by 18S rRNA gene sequencing from Pure Isolates |
MBS414 |
Next generation sequencing
Whole genome sequencing applications
Whole Genome Sequencing (WGS) has facilitated the thorough analysis of the complete genomic sequences of various organisms. This data is essential for gaining insights into the functional and evolutionary backgrounds of these organisms, as well as for detecting mutations and genetic variations linked to genetic disorders, cancer progression, disease outbreaks, and the emergence of strains with new pathogenic characteristics.
HiGx360® generates all the information required to comprehend the activities of all expected genes present in the organism in detail using a read-to-annotation technique. Since assembled genomes might not be entirely complete, one of the several long read sequencing methods can be used to fill in additional gaps.
- Sequence close relatives of reference-organisms or entirely novel organisms (de-novo)
- Obtain high-resolution and precise genomic characterization utilizing 2×150 or 2× 300 bp readings
- Data supplied with interactive QC report
- Get personalized bioinformatics analysis, including raw sequence reads, assembled contigs, and scaffolds
- Choose from basic or advanced bioinformatics
Recommendations
Source: HiMedia Laboratories Pvt. Ltd.
| |
Resequencing |
De-novo Sequencing |
| Depth/Coverage |
50X - 100X |
>100X |
| Read length chemistry |
2x75 to 2x300 |
| Sample Requirements |
Minimum quantity 0.5 μg, Minimum concentration = 20 ng/μl (Qubit), A260/280 = 1.8 - 2.0 |
Deliverables
- Run information, Statistics of raw data & FASTQ file
- Reference Genome Mapping files/ De-novo Genome Mapping files, Functional Annotations, Gene predictions, Gene annotations, Pangenome analysis
- Advanced analysis customized to the project needs
WGS services
Source: HiMedia Laboratories Pvt. Ltd.
| Description |
Service Codes |
| Whole Genome Sequencing (0.5 Gb Data) |
MBS201 |
| Whole Genome Sequencing (1 Gb Data) |
MBS201-1G |
| Whole Genome Sequencing (2 Gb Data) |
MBS201-2G |
| Whole Genome Sequencing (4 Gb Data) |
MBS201-4G |
| Whole Genome Sequencing (6 Gb Data) |
MBS201-6G |
| Whole Genome Sequencing (8 Gb Data) |
MBS201-8G |
| Whole Genome Sequencing (10 Gb Data) |
MBS201-10G |
| Whole Genome Sequencing (12 Gb Data) |
MBS201-12G |
| Whole Genome Sequencing (16 Gb Data) |
MBS201-16G |
| Whole Genome Sequencing (20 Gb Data) |
MBS201-20G |
| Whole Genome Sequencing (25 Gb data) |
MBS201-25G |
| Whole Genome Sequencing (30 Gb data) |
MBS201-30G |
| Whole Genome Sequencing (50 Gb data) |
MBS201-50G |
| Whole Genome Sequencing (60 Gb data) |
MBS201-60G |
| Whole Genome Sequencing (100 Gb data) |
MBS201-100G |
| Whole Genome Sequencing (30 Mb data, eg., Mycoplasma @50X) |
MBS201-30M |
| Whole Genome Sequencing (250 Mb data, eg., Bacteria @50X) |
MBS201-250M |
| Whole Genome Sequencing (Virus) (0.5GB) |
MBS202 |
| Whole Genome Sequencing (Virus, 10Kb @ 100X) |
MBS202-1K |
| Whole Genome Sequencing (Virus, 20Kb @ 100X) |
MBS202-2K |
| Whole Genome Sequencing (Virus, 50Kb @ 100X) |
MBS202-5K |
| COVIDSeq on MiSeq (per sample for 95 samples) |
MBS222 |
* Gb - Gigabases, Mb-Millionbases, KB - Kilobases
Whole exome sequencing applications
The human genome comprises 180,000 coding regions, representing 1.7% of its total structure. Approximately 85% of genetic-based human diseases arise from anomalies within these coding regions. The focused sequencing of these genomic areas is referred to as whole exome sequencing (WES). Compared to sequencing the entire human genome, exome sequencing offers a more economical alternative.
This sequencing technique employs a targeted capture strategy, utilizing biotinylated probes that are directed toward the coding regions of the fragmented human genome. These probes facilitate the capture and isolation of specific genomic segments using streptavidin-coated beads. The resulting fragments are then sequenced using the NextSeq™ 2000 platform. This approach achieves a significant enhancement in coverage, providing a 100-fold increase in the relevant areas of the genome.
- More affordable and widely accessible
- Sequencing coverage on target upto 30×
- Finding coding SNP variations with the same level of sensitivity as whole-genome sequencing
- Compared to whole-genome sequencing, a smaller data set allows for quicker and simpler analysis
- Standard sequencing coverage ≥ 50×; cancer sample ≥ 100×. More SNPs can be gained by increasing the coverage
- Exome enrichment options and affordable library preparation
Exome sequencing recommendations
Source: HiMedia Laboratories Pvt. Ltd.
| |
Whole Exome Sequencing |
Cancer Specific and/or Rare Variant Detection |
| Depth/Coverage |
100-300X |
500–1000X |
| Read length chemistry |
2x75 to 2x150 |
| Capture Region |
- 34 Mb target region
- 415,115 oligonucleotide probes
- 19433 genes
|
| Sample Requirements |
Required quantity 1.0 – 0.5 μg, Minimum concentration = 20 ng/μl (Qubit), A260/280 = 1.8 - 2.0 |
Deliverables
- SNPs and InDels calling, Variant annotation, SNVs concordance, Tumor-Normal paired analysis
- Run information, Statistics of raw data & FASTQ/VCF files
- Mapping statistics, Statistics of sequencing reads
WES services
Source: HiMedia Laboratories Pvt. Ltd.
| Description |
Service Codes |
| Whole Exome Sequencing |
MBS212-45M |
| Whole Exome Sequencing |
MBS212-64M |
* Gb - Gigabases, Mb-Millionbases, KB - Kilobases
Metagenomics and microbiome applications
Life on Earth depends on microorganisms. The functional potential of microbial communities can be inferred from descriptive metrics such as the metagenome, which is the collection of genes and genomes of the microbiota, and the microbiota, which is the collection of microorganisms existing in a certain environment.
A more thorough knowledge of hypothesis testing is made possible by microbiome interactions, which relate to the overall habitat, including the microbes, their genomes, and the surrounding environmental factors. The Hi-Gx360® NGS service allows users to:
- Sequence a set of microbiota genes or genomes in any sample
- Sequence the 16S and ITS marker, as well as additional genomic regions and marker genes
- Learn about the functional importance of microbial diversity in detail
- Use in-depth sequencing to find microbial species that are scarce
Metagenome sequencing
Shotgun metagenomics is a reliable method for studying environmental, agricultural, and human microbiomes, identifying and monitoring pathogens, and tracking antimicrobial resistance genes. Some of the most popular uses are:
- Discovery of novel enzymes and metabolic pathways
- Relative abundance analysis of microbial communities
- Identification and classification of microbial communities
- Novel biomarker discoveries
- De-novo assembly and characterizations of novel genomes
Amplicon-based microbiome sequencing
- A promising method for high-throughput phylogenetic study of communities of microbes
- Based on phylogenetic marker genes: 16S rRNA genes, and ITS region
- Enables the characterization of taxonomic diversity and species identification.
- Provides detailed insight into the genus and species
Advantages of amplicon-based phylogenetic gene marker sequencing
- Marker genes are omnipresent
- Marker genes are relatively abundant compared to other genes
- Measures phylogenetic connections between various taxa
- Easily determine the relative abundance of microbial communities
- Highly cost-effective
Recommendations
Source: HiMedia Laboratories Pvt. Ltd.
| |
Shotgun Metagenome |
Amplicon-based MiCrobiome |
| Objective |
- Discovery metagenomics
- Relative abundance of microbial communities
- Metagenome assembled genome sequencing
|
Taxonomic profiling |
| Reads per sample |
25-500 Million paired end reads* |
Over 1 Lakh paired end reads* |
| Read length chemistry |
1x150 to 2x300 |
| Sample Requirements |
Minimum Quantity 1 μg, Minimum Concentration = 50 ng/μl (Qubit), OD 260/280=1.8-2.0 |
Deliverables
- Run information, Statistics of raw data and FASTQ files
- Taxonomic profiles, Functional Annotations, Metagenome assembled genomes (MAGs)
- Advanced analysis customized to the project needs
Shotgun-metagenome based services
Source: HiMedia Laboratories Pvt. Ltd.
| Description |
|
Service Codes |
| Metagenome sequencing (soil/ sediment/water) |
(5 Gb Data) |
MBS213-5G |
| (10 Gb Data) |
MBS213-10G |
| (15 Gb Data) |
MBS213-15G |
| (20 Gb Data) |
MBS213-20G |
| Metagenome sequencing (Clinical) |
(1 Gb Data) |
MBS214-1G |
| (5 Gb Data) |
MBS214-5G |
| (10 Gb Data) |
MBS214-10G |
| (15 Gb Data) |
MBS214-15G |
| (20 Gb Data) |
MBS214-20G |
| Metagenome sequencing Environmental DNA (eDNA) |
(5 Gb Data) |
MBS215-5G |
| (10 Gb Data) |
MBS215-10G |
| (15 Gb Data) |
MBS215-15G |
| (20 Gb Data) |
MBS215-20G |
* Gb - Gigabases, Mb-Millionbases, KB - Kilobases
Amplicon-based microbiome services
Source: HiMedia Laboratories Pvt. Ltd.
| Description |
|
Service Codes |
| Microbiome sequencing from Community DNA (16S) |
(2.5 lakh PE reads) |
MBS203-2L |
| (5 lakh PE reads) |
MBS203-5L |
| (10 lakh PE reads) |
MBS203-10L |
| Microbiome sequencing from Insect/Food sample (16S) |
(2.5 lakh PE reads) |
MBS204-2L |
| (5 lakh PE reads) |
MBS204-5L |
| Microbiome sequencing from Swabs/body fluids (16S) |
(2.5 lakh PE reads) |
MBS205-2L |
| (5 lakh PE reads) |
MBS205-5L |
| Microbiome sequencing from Soil/Sediment/Water/Fecal (16S) |
(2.5 lakh PE reads) |
MBS206-2L |
| (5 lakh PE reads) |
MBS206-5L |
| Microbiome sequencing from Community DNA (ITS) |
(2.5 lakh PE reads) |
MBS207-2L |
| (5 lakh PE reads) |
MBS207-5L |
| Microbiome sequencing from Insect/Food sample (ITS) |
(2.5 lakh PE reads) |
MBS208-2L |
| (5 lakh PE reads) |
MBS208-5L |
| Microbiome sequencing from Swabs/body fluids (ITS) |
(2.5 lakh PE reads) |
MBS209-2L |
| (5 lakh PE reads) |
MBS209-5L |
| Microbiome sequencing from Soil/Sediment/Water/Fecal (ITS) |
(2.5 lakh PE reads) |
MBS210-2L |
| (5 lakh PE reads) |
MBS210-5L |
| 16S Microbiome ready-to-load |
(50,000 PE reads) |
MBS216 |
| (1 lakh PE reads) |
MBS216-1L |
| (2.5 lakh PE reads) |
MBS216-2L |
| (5 lakh PE reads) |
MBS216-5L |
Supplemental services
Only available in conjunction with other service codes, not independently.
Source: HiMedia Laboratories Pvt. Ltd.
| Description |
Service Codes |
| DNA Fragment QC |
MBS304 |
| RNA QC with RIN value |
MBS306 |
Transcriptomics
RNA sequencing (RNA-seq) is a potent and frequently used tool for transcriptome research, revolutionizing molecular biology. Researchers may analyze gene expression in unprecedented depth, gaining vital insights into biological processes and disease pathogenesis. RNA-seq converts RNA molecules into cDNA fragments, which are subsequently sequenced utilizing high-throughput technology.
- Identify post-transcriptional modifications, including RNA editing
- Identify and measure gene expression levels, locate unique transcripts, and examine alternative splicing patterns
- Wide dynamic range and low input requirements
- Work with a range of sample types, including single cells and complex tissues
- Integrate with other omics data to get a more complete picture of biological systems
RNA sequencing services
Source: HiMedia Laboratories Pvt. Ltd.
| Description |
Service Codes |
| de novo RNA Sequencing/Transcriptome Sequencing (10M PE reads, 2x150) |
MBS230-10M |
| de novo RNA Sequencing/Transcriptome Sequencing (20M PE reads, 2x150) |
MBS230-20M |
| de novo RNA Sequencing/Transcriptome Sequencing (30M PE reads, 2x150) |
MBS230-30M |
| Top Up Sequencing (10M PE reads, 2x150) |
MBS230-10MTUP |
| RNA Sequencing/Transcriptome Sequencing, 10Gb data(W~67M PE reads, 2x150) |
MBS230-10G |
| mRNA Sequencing/RNA Sequencing/Whole Transcriptome Sequencing (10M PE reads, 2x75) |
MBS231-10M |
| mRNA Sequencing/RNA Sequencing/Whole Transcriptome Sequencing (20M PE reads, 2x75) |
MBS231-20M |
| mRNA Sequencing/RNA Sequencing/Whole Transcriptome Sequencing (30M PE reads, 2x75) |
MBS231-30M |
Custom sequencing

Image Credit: HiMedia Laboratories Pvt. Ltd.
Sequencing services
Source: HiMedia Laboratories Pvt. Ltd.
| Category |
Description |
| NGS |
16S Microbiome ready-to-load |
| NGS |
MiSeq ready-to-load |
| NGS |
Custom Sequencing on MiSeq |
| NGS |
NextSeq ready-to-load |
| NGS |
NextSeq ready-to-run |
Bioinformatics services
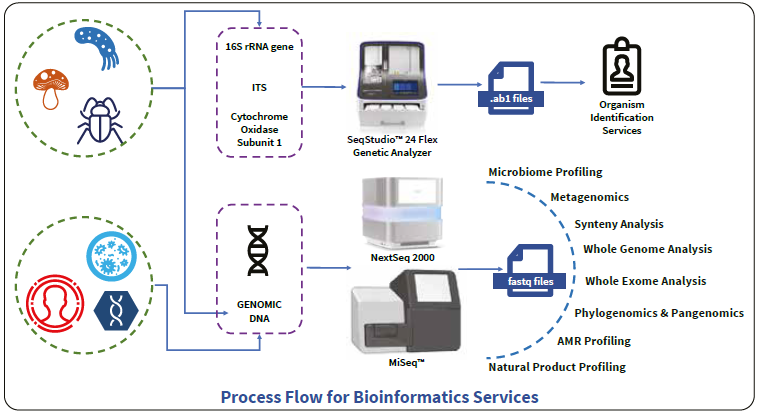
Image Credit: HiMedia Laboratories Pvt. Ltd.
Analyzing both small and large sequence datasets from Next-Generation Sequencing (NGS) presents a significant challenge, especially with massively parallel sequencing. This is primarily due to limitations in scalable computational power and insufficient understanding of the inner workings of various bioinformatics (BioIT) tools. Furthermore, data privacy remains a daunting hurdle for researchers handling patient data.
One of the main difficulties, particularly in massively parallel sequencing, is analyzing customers’ NGS files, regardless of how big or little they are. The primary causes of issues are a lack of scalable computing capacity and an ignorance of the inner workings of various bioinformatic (BioIT) technologies.
For academics dealing with patient data, data privacy is still a formidable obstacle. Each of these issues has been methodically resolved by Hi-Gx360® at our cutting-edge bioinformatics data center. This comprises:
- Secure datasets that are never made publicly available online
- Open and transparent exchange of information on the analytical process
- Analysis was performed using highly scalable and tested open-source packages
- Secure interactive reports that are comprehensive and information-rich
- Publication grade data visualizations
- Direct Consultations with the highly qualified scientists on project design
With the bioinformatics services, users can accurately achieve the following:
- Combine with short and long reads libraries to obtain complete genome sequence
- Identify repetitive regions, structural variants and complex rearrangements for de-novo assemblies
- Identify organisms with confidence
- Assemble, annotate and analyze whole genomes
- Locate low frequency mutations & identify InDels
- Uncover genetic traits, such as antibiotic resistance markers, etc.
Bioinformatics services
BioIT services for Sanger data
Source: HiMedia Laboratories Pvt. Ltd.
| Description |
Service Codes |
| Phylogenetic Analysis (16S/18S/ITS/any other gene based) |
MBS020 |
| NCBI submissions |
MBS021 |
| Primer Designing |
MBS024 |
| Custom Analysis for Sanger Data |
MBS029 |
BioIT services for NGS data
Source: HiMedia Laboratories Pvt. Ltd.
| Description |
Service Codes |
| Whole Genome analysis for Bacteria/ Archaea (Basic) |
MBS001 |
| Whole Genome analysis for Bacteria/ Archaea (Advanced) |
MBS002 |
| Whole Genome analysis for Fungi (Basic) |
MBS003 |
| Whole Genome analysis for Fungi (Advanced) |
MBS004 |
| Whole Genome analysis for Virus |
MBS005 |
| Genome Mining for Natural Products from Bacteria/Archaea (Requires MBS001) |
MBS010 |
| Genome Mining for AMR and other features from Bacteria/Archae (Requires MBS001) |
MBS011 |
| Genome Based Microbial identification (Requires MBS001) |
MBS012 |
| Microbiome profiling (Basic Analysis 16S/18S/ITS/any other gene based) |
MBS013 |
| Microbiome profiling (Advanced Analysis 16S/18S/ITS/any other gene based) |
MBS014 |
| Shotgun Metagenomics (Bacterial/ Archaeal/Viral communities) |
MBS015 |
| Genome resolved Metagenomics (Bacterial/Archaeal/Viral communities) |
MBS016 |
| Transcriptome analysis (Kallisto/Salmon or prokaryote) |
MBS017 |
| Transcriptome analysis (HiSat/Star or eukaryote) |
MBS018 |
| Whole Genome analysis for Yeast (Basic) |
MBS022 |
| Whole Genome analysis for Yeast (Advanced) |
MBS023 |
| SRA Submission of NGS Dataset |
MBS025 |
| Whole Exome Analysis (Human) |
MBS027 |
| Mitochondrial Whole Genome Analysis |
MBS028 |
| Custom Analysis for NGS Data |
MBS026 |
Specialized in-silico genome mining services
Natural product discovery
Some of the best chemists in human history are microorganisms, which also generate a variety of bioactive chemicals. Even before beginning the expensive process of chemical characterization, genome mining can help forecast the organism's capacity for biosynthesis. The journey from discovery to manufacturing can be shortened by using the information gathered from genome mining to inform chemical synthesis.
MBS010 Genome Mining for Natural Products from Bacteria Archaea (requires MBS001)
Antimicrobial resistance profile
Antimicrobial resistance (AMR) poses a global risk to human health and development. AMR is one of the top 10 worldwide public health problems, according to WHO. Identifying control and preventative techniques is crucial for combating the growing threat of AMR. Genome mining is an effective method for accurately predicting AMR in bacteria. Hi-Gx360® offers assistance in identifying AMR indicators from an organism's genomic sequence.
MBS011 Genome Mining for AMR and other features from Bacteria/Archaea (Requires MBS001).
Products of Hi-Gx360
Hi-Gx360® library preparation kits
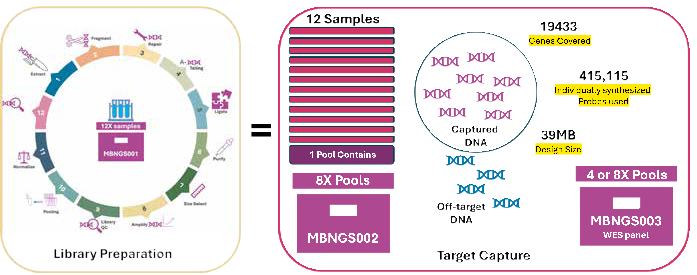
Whole Genome and Whole Exome Workflow. Image Credit: HiMedia Laboratories Pvt. Ltd.
NGS library preparation is an important stage in sequencing operations. This approach enables researchers to examine a variety of biological topics, including genome sequencing and metagenomics, with high accuracy and efficiency. The Hi-Gx360® Library Preparation Kits can efficiently prepare libraries from many DNA sources, including human, plant, animal, and microorganisms.
Source: HiMedia Laboratories Pvt. Ltd.
| Category |
Description |
Service Codes |
| NGS |
Hi-Gx360® DNA Library Preparation Kit for Illumina® Sequencing Platform |
MBNGS001 |
| NGS |
Hi-Gx360® Targeted Capture & Enrichment of DNA Libraries |
MBNGS002 |
| NGS |
Hi-Gx360® WES Panel |
MBNGS003 |
Solution
Cost per test
A single library prep kit can be used for many workflows, resulting in a cost-effective test solution.
Customers
The variety of pack sizes can help small, medium, and large-scale end users.
Sustainability
Modularly increase pack sizes to avoid kits from expiring too quickly.
Multipurpose
One core library prep kit for multiple applications.
Product benefits
Using a reproducible library preparation and target capture technique ensures reliable sequencing data for users while allowing for customization of gene panels.
DNA library profile
Consistent library profiles across sample types
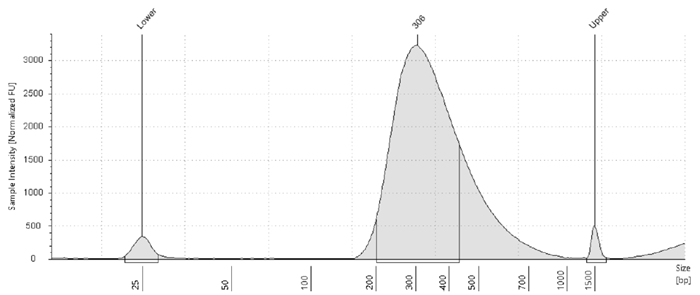
Illumina library profile of Blood DNA. Image Credit: HiMedia Laboratories Pvt. Ltd.
Product overview
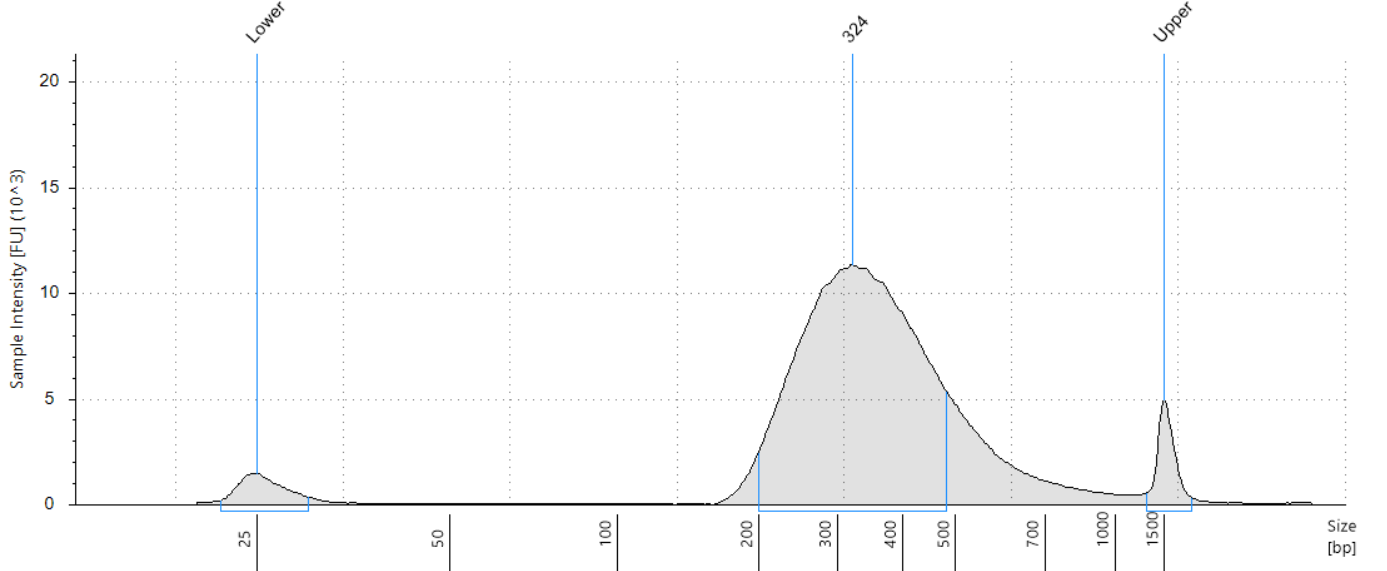
Illumina library profile of Bacterial DNA. Image Credit: HiMedia Laboratories Pvt. Ltd.
Excellent read quality in 2× 150 base chemistry
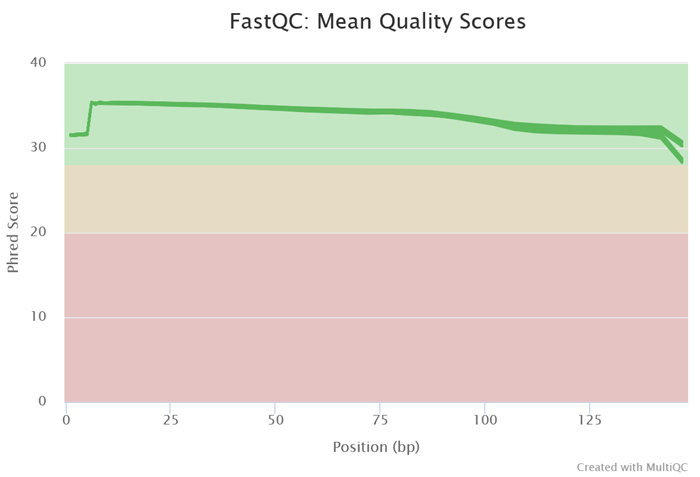
Image Credit: HiMedia Laboratories Pvt. Ltd.
Product benefits
Using a reproducible library preparation and target capture technique ensures reliable sequencing data for users while allowing for customization of gene panels.
Consumables for Sequencing
Library preparation kits
Source: HiMedia Laboratories Pvt. Ltd.
| Category |
Description |
Service Codes |
| NGS |
Hi-Gx360® DNA Library Preparation Kit for Illumina® Sequencing Platform |
MBNGS001 |
| NGS |
Hi-Gx360® Targeted Capture & Enrichment of DNA Libraries |
MBNGS002 |
| NGS |
Hi-Gx360® WES Panel |
MBNGS003 |
Extraction kits
Source: HiMedia Laboratories Pvt. Ltd.
| Product Name |
Product Code |
| HiPurA® Blood Genomic DNA Miniprep Purification Kit |
MB504 |
| HiPurA® Bacterial Genomic DNA Purification Kit |
MB505 |
| HiPurA® Soil DNA Purification Kit |
MB542 |
HiPurA® HP Fungal DNA Purification Kit
(following bead beating method) |
MB576 |
| HiPurA® Pre-filled Cartridges for Bacterial DNA Purification |
MB505PC16-48PR |
| HiPurA® Pre-filled Plates for Bacterial DNA Purification |
MB505MPF16-96PR |
| HiPurA® Pre-filled Cartridges for Fungal DNA Extraction |
MB576PC16-48PR |
| HiPurA® Pre-filled Plates for Fungal DNA Extraction |
MB576MPF16-96PR |
Reagents
Source: HiMedia Laboratories Pvt. Ltd.
| Product Name |
Product Code |
| 1kb DNA Ladder |
MBT051 |
| 50bp DNA Ladder |
MBT084 |
| 100bp DNA Ladder |
MBT049 |
| 6X Gel Loading Buffer |
ML015 |
| 2X PCR TaqMixture |
MBT061 |
| 2-Propanol |
MB063 |
| Poly(ethylene glycol) MW 8000 |
MB150 |
| 1N Sodium hydroxide |
ML195 |
| 5M Sodium chloride |
ML008 |
| 1M Tris-Cl, pH 8.5 |
ML152 |
| 3M Sodium acetate, pH 5.2-5.4 |
ML009 |
| 0.5M EDTA, pH 8.0 |
ML014 |
| Molecular Biology Grade Water for PCR |
ML065 |
| RNase Kil™ |
ML162 |
| 1X PBS Solution |
ML116 |
| Agarose, Ultrapure, Low EEO |
MB229 |
| HiPurA® Mag Beads for Cleanup |
ML239 |
| DNA Kil™ |
ML221 |
Lab Consumables
Source: HiMedia Laboratories Pvt. Ltd.
| Product Name |
Product Code |
| PCR Blocks (Full-skirt, natural, sterile, DNA/RNAse free 96 well, 0.1ml) |
PR2 |
| PCR Blocks (Semi-skirt, natural, sterile, DNA/RNAse free 96 well, 0.2 ml volume) |
PR3 |
| PCR Blocks (Non-skirt, natural, sterile, RNase, DNase free, 96 well, 0.2 ml volume) |
PR19 |
| Aluminium Sealing Film |
PR20 |
| Optical Sealing Film 96 well PCR plate |
PR18 |
| 96 Well Plate for Sequencing/PCR |
PR26 |
| Silicone Mat for 96- well plates (PCR) |
PR27 |
| 8 strip PCR tubes with optically clear attached flat caps for Real-time PCR |
PR23 |
| PCR Tubes, Flat lid Autoclavable, Conical Bottom, with Graduation |
CG281 |
PCR Tubes, Thin walled DNase/RNase free,
Autoclavable, Conical Bottom, without Graduation, Flat lid |
PW1255 |
| Micro Centrifuge Tube - B |
PW146 |
| HiPer® Lock MicroCentrifuge Tube, 2.0 ml |
MBLA017 |
| Barrier Tips, 10 μl |
LA749A |
| Barrier Tips XL, Max. capacity 10 μl |
LA749XL |
| Barrier Tips, 20 µl |
LA750A |
| Barrier Tips XL, Max Capacity 20 µl |
LA750XL |
| Barrier Tips, 100 μl |
LA1104A |
| Barrier Tips, 200 µl |
LA751A |
| Barrier Tips, 1000 μl |
LA859A |
| Polypropylene Cryogenic Storage Box |
PW1215 |