Proteins are large biological molecules that are integral to complex networks. These networks control key physiological processes via signaling pathways and feedback. Such regulatory functions largely depend on the ability of proteins to target and form specific non-covalent complexes with other proteins. With each interaction, the protein structural changes occur through specific electrostatic and van der Waals interactions, the breaking and formation of hydrogen bonds and the burial of surfaces exposed to solvents.
It is estimated that each protein in an organism interacts with an average of five other proteins (Piehler, 2005) and based on its concentration or location in the cell, the same protein can form different interactions (Liddington, 2004). In addition to these intermolecular interactions, interactions between proteins also take place at the intra-molecular level, at interfaces between subunits within a protein and between domains.
This article explores the application of isothermal titration calorimetry (ITC) for quantitative determination of the thermodynamic properties that power interactions between proteins. It also elucidates how ITC can be utilized to measure protonation/de-protonation events that occur upon binding. However, using ITC, only one component is titrated into another and this technique is generally limited to analyzing bimolecular interactions.
Analyzing Protein-Protein Interactions
For interactions of two protein surfaces to occur, good hydrophobic, charge and shape complementarity are required to ensure the resulting interface is as well packed as within the protein. Given this requirement, along with prior knowledge about which proteins interact (by utilizing yeast two hydroid systems, protein fragment complementation assays, or a combination of both these methods, for example), it should be possible theoretically to predict the interfaces though which two proteins interact, along with the binding mechanism. However, in practice, dynamic events that take place in both the protein and the solvent make this prediction complicated.
The inter- or intramolecular interaction of two protein moieties often gives rise to significant structural variations both in the vicinity of the interacting interfaces and also distant from the contact site. This results in modified hydrogen bonds, electrostatic and hydrophobic interactions and substantial reorganization of bound water molecules. Interaction between two protein interfaces will result in unique changes to the protein structure and a unique rearrangement of water molecules distributed between the bound and free states. These rearrangements of solvents result in enthalpic and entropic changes to the system.
ITC represents a direct and quantitative method for defining the thermodynamic properties of interactions between proteins. Although other methods allow measurement of the stoichiometry and binding affinity, the ITC technique is more direct. For instance, analytical ultracentrifugation takes a significant amount of time, surface plasmon resonance requires immobilization that can disrupt binding, and spectroscopic analyses often involve the use of labels. Moreover, these methods do not enable the enthalpy of binding to be directly measured and this has to be determined from the van't Hoff relationship. By contrast, ITC precisely determines the heat evolved during the interaction of two protein interfaces and provides the stoichiometry (n), binding constant (Ka) as well as the enthalpy of binding (AH). This technique also enables the change in entropy, ∆S, and the change in Gibbs energy of the system, ∆G, to be acquired from:
- RTIn K = ∆G = ∆H - T∆S
Where T represents the absolute temperature and R is the universal gas constant. Given that entropic and enthalpic contributions to the complex’s stability reflect different types of interactions, each protein complex will react differently to mutational and environmental changes, thereby allowing the relative importance of these interactions to be assessed.
It is possible to estimate the pKa and enthalpy of ionization of each ionizable group involved in the binding reaction by performing the titration in several buffers with different enthalpies of ionization but the same pH. In addition, by carrying out the experiment at two temperatures (T1 and T2), the variation in heat capacity of the system, AC, can be acquired from:
∆Cp - (∆Hn - ∆HT2) /(T2 - Tx).
Practical Considerations: Types of Protein-protein Interactions Compatible with ITC
Three types of protein-protein interactions exist and include homomeric, heteromeric, and domain-domain interactions. In domain-domain interactions, two domains which are separately folded form an interface, typically involving residues widely separated in sequence. Since domain-domain interactions take place inside the same polypeptide chain, ITC experiments cannot be used to study them, unless protein fragments are used. As an alternative, such interactions are often probed by mutating important residues believed to be key to domain interactions and ascertaining the stability of the mutants through differential scanning calorimetry (DSC). The ITC technique is perfect for analyzing heterodimeric interactions. A solution containing one protein is incrementally injected into a solution of the second protein. The binding of the two proteins with each addition of titrant emits heat which is proportional to the concentration of the complex after injection:
qi =V∆Ha ([M1M2] I - [M1M2]I -1)
Where V is the volume of the cell, qi is the heat produced by the ith injection, [M1M2]I is the concentration of the complex following the ith injection, and ∆Ha is the enthalpy of binding. Titrant (M1) is introduced to the second protein (M2) until it is saturated and binding is no longer observed. As the concentrations of both proteins are known during the course of titration, nonlinear regression analysis of qi allows the binding constant (Ka = [M1M2] /[M1]M2]), ∆Ha and ∆S to be calculated.
The ITC method is also suitable for examining the dissociation of homodimers. In the calorimetric cell, the dimeric protein is titrated into buffer and dilution of the protein leads to dissociation into its constituent subunits. The dissociation constant Kd establishes the distribution of the subunits between the dimeric (M2) and monomeric (M) states {Kd - [M]2[M2]). Given that the heat associated with each protein injection is proportional to the monomer concentration following the ith injection, the enthalpy due to the protein’s dissociation into monomers (∆Hd) can be determined as follows:
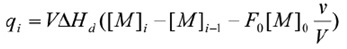
Where v is the injection volume, V is the volume of the calorimetric cell, F0, is the fraction of monomer in the dimer solution in the syringe and [M]0 is the concentration of protein in the cell. As the titration takes place and the concentration of the monomer in the cell rises, the dissociation reaction becomes increasingly less favorable until additional injections of dimer do not result in any significant heat production. It is then possible to determine the enthalpy due to dissociation, the dissociation constant, and the entropy, through nonlinear regression analysis.
Concentration Ranges Compatible with ITC
Equilibrium processes involve the reversible inter-conversion of two components, meaning experiments have to be carried out at suitable concentrations so that any variation in the concentration of the first component results in quantifiable change in the concentration of the second component. During binding experiments wherein interactions occur between two proteins, concentrations must be selected so that during the first third of the titration, all the titrant binds to the macromolecule in the cell, and during the last third of the titration, the macromolecule is saturated with titrant and little or no additional binding occurs.
In cases where precise determination of the enthalpy of binding is most important, then the titrant protein must be introduced into a large surplus of the binding protein in the sample cell. While it is not possible to determine the stoichiometry of binding or binding constant from this experiment, the binding heat can be precisely measured after subtraction of a blank run designed to compensate for buffer mismatch, dilution effects and so on.
Practical Example: Characterization of Hetero-Dimer Formation
The ITC technique used for defining heterodimer formation is shown by the example of soybean trypsin inhibitor binding to porcine pancreatic trypsin.
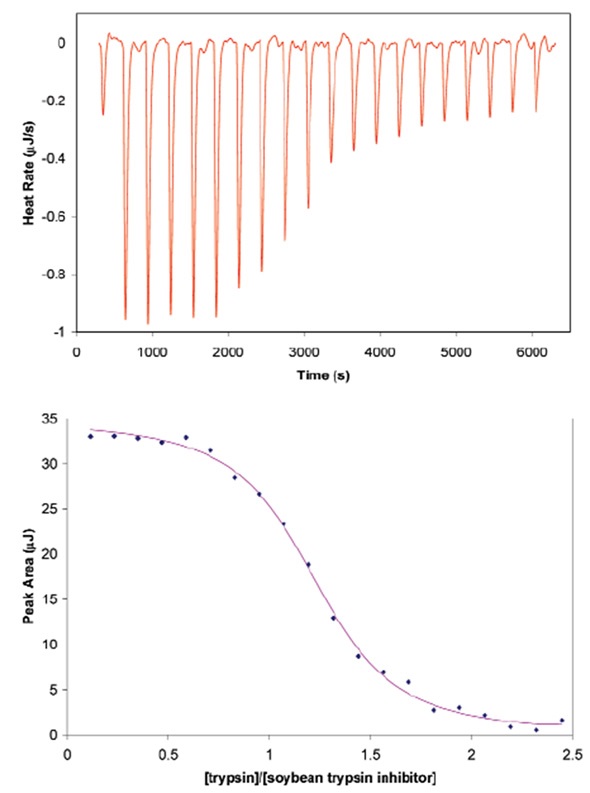
Figure 1. Titration of porcine pancreatic trypsin into soybean trypsin inhibitor using a CSC model 5300 ITC-iil. Both proteins were dialyzed at 4°C against 25mM potassium acetate pH 4.5 buffer containing 10mM calcium chloride. Soybean trypsin inhibitor (2.1µM) was loaded in the 1.0mL sample cell and trypsin (440µM) was loaded in the 100µL syringe. Twenty, 5µL aliquots of ligand were titrated into the sample cell while the temperature of the system was maintained at 25°C. Top panel: The signal (heat) produced following each addition of protein to the inhibitor. Bottom panel: Integration of the heats over the time course of the experiment; the µJ in each peak are plotted against the mole ratio of the titrant (trypsin) to inhibitor (soybean trypsin inhibitor). The inhibitor was placed in the sample cell rather than the syringe due to its low solubility. Ka of binding: 1.4 x 1O7; stoichiometry: 1.15; enthalpy of binding: 135KJ mol-1.
To achieve a titration curve with adequate points for precise curve fitting, titrant should be added incrementally until it exceeds stoichiometric binding to the second protein. This experiment of trypsin and inhibitor binding was designed using the data simulation module Experiment Design Wizard, which is included in all CSC ITC equipment. This module enables experimental parameters such as predicted thermodynamic results and concentration to be inputs and outputs a simulation of the predicted experimental thermogram. The module also ensures that sample material and time and are not wasted on runs that are not able to produce useable data owing to inaccurate concentration options. Figure 1 shows the conditions used to establish the binding enthalpy, the stoichiometry of binding, and the association constant for soybean trypsin inhibitor binding to trypsin.
Protonation Effects
When binding occurs between two protein interfaces, it is possible to change the pKa values of ionizable groups at the interfaces due to electrostatic or solvation changes triggered by the binding event. This leads to deprotonation or protonation of these residues. When this takes place, the binding enthalpy will depend on the ionization enthalpy of the buffer (∆Hbuf) and the association constant will depend on pH. A series of ITC experiments can be carried out to ascertain whether a protonation or deprotonation reaction is taking place and, if so, the number of protons involved.
The ITC titrations are carried out in buffers with varying ionization enthalpies. The quantified enthalpy, ∆Happ is equal to the enthalpy of binding (∆Hbind, which is pH dependent but independent of the buffer used), to the number of protons (nH,) released or adsorbed by the binding reaction and to the ionization enthalpy of the buffer:
∆app = ∆Hbmi + nH∆Hbuf
A plot of ∆Hjpp against the buffer ionization enthalpy will give a straight line, making it possible to estimate the nH from the slope and ∆Hbind from the intercept with the y axis. There is no net transfer of protons if the slope is zero. If nH is negative, protons are transferred from the protein complex to the buffer solvent and if nH is positive, protons are transferred from the solvent to the protein complex. This approach can be used to calculate the enthalpy of ionization and pKa of each ionizable group involved in the protonation or deprotonation reaction.
Conclusion
Interactions between proteins regulate all physiological processes and are implicated in many disease conditions. As a result, more and more studies have been performed to gain insight into the thermodynamics driving certain binding interactions, with the intent that the disruption or selective control of these interactions may help explain important biological mechanisms.
ITC is a simple and general approach that can be used to establish a complete thermodynamic description of the specific binding interactions between two proteins, in addition to the association constant and stoichiometry of binding. Furthermore, if structural information on the binding interface is available, it is possible to identify the contributions of specific amino acids regulating the binding event and their thermodynamic contributions.
Acknowledgement
Created from articles authored by Christin T. Choma, TA Instruments, 109 Lukens Drive, New Castle, DE 19720, USA.
Sources and Further Reading
(Preference has been given to current references. Citation does not imply that a paper is necessarily the original reference to a study.)
- Baker, B. M. and K. P. Murphy. (1996) Evaluation of linked protonation effects in protein binding reactions using isothermal titration calorimetry. Biophys. J.71,2049-2055.
- Baker, B. M. and K. P. Murphy. (1997) Dissecting the energetics of a protein-protein interaction: the binding of ovomucoid third domain to elastase. J. Bioi. Chem.268, 557-569.
- Barbieri, C. M. and D. S. Pilch. (2006) Complete thermodynamic characterization of the multiple protonation equilibria of the aminoglycoside antibiotic paromomycin: a calorimetric and natural abundance 15N NMR study. Biophys. J. 90, 13381349.
- Christensen, J. J., L. D. Hansen and R. M. Izatt. (1976). Handbook of proton ionization heats and related thermodynamic quantities. Wiley, New York.
- Christensen, T., D. M. Goodin, J. E. King and E. J. Toone. 2003 Additivity and the physical basis of multivalency effects: a thermodynamic investigation of the calcium EDTA interaction, JACS 125, 7357-7366.
- Ciaccio, C. et al. (2004) Proton linkage for CO binding and redox properties of bovine lactoperoxidase. Biophys. J. 86,448-454.
- Crnogorac, M. M., G. M. Ullmann and N. M. Kostic. (2001) Effects of pH on protein association: Modification of the protein-linkage model and experimental verification of the modified model in the case of cytochrome c and plastocyanin. JACS 123,10789-10798.
- Doyle, M. L. (1997) Characterization of binding interactions by isothermal titration calorimetry. Curr. Opin. Biotechnol. 8, 31-35.
- Doyle, M. L., G. Louie, P. R. Dal Monte and T. K. Sokoloski. (1995) Tight binding affinities determined from thermodynamic linkage to protons by titration calorimetry, Meth. Enzymol. 259,183-194.
- Doyle, M. L. and P. Hensley. (1998) Tight ligand binding affinities determined from thermodynamic linkage to temperature by titration calorimetry. Meth. Enzymol.295,88-99.
- Frisch, C, G. Schreiber, C. M. Johnson and A. R. Fersht. (1997) Thermodynamics of the interaction of Barnase and Barstar: changes in free energy versus changes in enthalpy on mutation. J. Biol. Chem. 267,696-706.
- Gomez, J. and E, Freire. (1995) Thermodynamic mapping of the inhibitor site of the aspartic protease endothiapepsin. J. Mol. Biol. 252, 337-350.
- Hazbun, T. R. et al. (2003) Assigning function to yeast proteins by integration of technologies. Mol. Cell. 12,1353-1365.
- Hu, C. D. and T. K. Kerppola. (2003) Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21, 539-545.
- Jelesarov, I and H. R. Bosshard. (1994) Thermodynamics of ferredoxin binding to ferredoxin:NADP+ reductase and the role of water at the complex interface. Biochemistry 33,13321 -13328.
- Jelesarov, I. and H. R. Bosshard. (1999) Isothermal titration calorimetry and differential scanning calorimetry as complementary tools to investigate the energetics of biomolecular recognition. J. Mol. Recognit. 12, 3-10.
- Kaul, M., C. M. Barbieri, J. E. Kerrigan and D. S. Pilch. (2003) Coupling of drug protonation to the specific binding of aminoglycosides to the A site of 16 S rRNA: elucidation of the number of drug amino groups involved and their identities. J. Mol. Biol. 326,1373-1387.
- Ladbury, J. E. (1996) Just add water! The effect of water on the specificity of protein-ligand binding sites and its potential application to drug design. Curr. Opin. Struct. Biol. 3, 973-980.
- Ladbury, J. E. and B. Z. Chowdhry. (1996) Sensing the heat: the application of isothermal titration calorimety to thermoydynamic studies of biomolecular interactions. Chemistry & Biology 3, 791-801.
- Ladbury, J. E. and M. A. Williams. (2004) The extended interface: measuring non-local effects in biomolecular interactions. Curr. Opin. Struct. Biol. 14,562-569.
- Lakey, J. H. and E. M. Raggett. (1998) Measuring protein-protein interactions. Curr. Opin. Struct. Biol. 8,119-123.
- Liddington, R. C. (2004) Structural basis of proteinprotein interactions. In H. Fu (Ed.) Methods in Molecular Biology, vol. 261: Protein-protein interactions: methods and protocols, pp.3-13, Humana Press, Totowa, NJ,
- Lobo, B. A„ G. S. Koe, J. G. Koe and C. R. Middaugh. (2003) Thermodynamic analysis of binding and protonation in DOTAP:DOPE (1:1):DNA complexes using isothermal titration calorimetry. Biophys. J. 104,67-78.
- Nguyen, B., J. Stanek and W. D. Wilson. (2006) Binding-linked protonation of a DNA minor-groove agent. Biophys. J. 90,1319-1328.
- O'Brien, R„ J. E. Ladbury and B. Z. Chowdry. (2001) Isothermal titration calorimetry of biomolecules. p. 263-286. In S. E. Harding and B. Z. Chowdry (Eds.) Protein-Ligand Interactions: hydrodynamics and calorimetry. Oxford University Press, Oxford.
- Parker, M. H. et al. (1999) Analysis of the binding of hydroxamic acid and carboxylic acid inhibitors to the stromelysin-1 (matrix metalloproteinase-3) catalytic domain by isothermal titration calorimetry. Biochemistry 38,13592-13601.
- Piehler, J. (2005) New methodologies for measuring protein interactions in vivo and in vitro. Curr. Opin. Struct. Biol. 15,4-14.
- Pierce, M. M., C. S. Raman and B. T. Nail. (1999) Isothermal titration calorimetry of protein-protein interactions. Methods 19,213-221.
- Privalov, P. L. and A. I. Dragan. (2006) Microcalorimetry of biological macromolecules. Biophys. Chem. 122, 158-169.
- Raschke, T. M. (2006) Water structure and interactions with protein surfaces. Curr. Opin. Struct. Biol. 16, 152-159.
- Toby, G. G. and E. A. Golemis. (2001) Using the yeast interaction trap and other two-hybrid-based approaches to study protein-protein interactions. Methods 24,201-217.
- Velazquez-Campoy, A., I. Luque, M. J. Todd, M. Milutinovich, Y. Kiso and E. Freire. (2000) Thermodynamic dissection of the binding energetics of KNT-272, a potent HIV-1 protease inhibitor. Prot. Sci. 9,1801-1809.
- Velazquez-Campoy, A., S. A. Leavitt and E. Freire. (2004) Characterization of protein-protein interactions by isothermal titration calorimetry. In H. Fu (Ed.) Methods in Molecular Biology, vol. 261: Proteinprotein interactions: methods and protocols, pp. 35-54, Humana Press, Totowa, NJ.
- Velazquez-Campoy and E. Freire. (2005). ITC in the post-genomic era...? Priceless. Biophysical Chemistry 115,115-124.
- Zhao, L. and J. Chmielewski. (2005) Inhibiting proteinprotein interactions using designed molecules. Curr. Opin. Struct. Biol. 15,31 -34.
About TA Instruments
 TA Instruments' reputation for high technology products, quality manufacturing and unbeatable after sales support is why more customers recommend TA products to their colleagues around the world. Headquartered in New Castle, DE, TA Instruments prides ourselves in the technical competence and professionalism that our sales force offers. We are the only thermal analysis, rheology, and microcalorimetry supplier recognized worldwide for our prompt, courteous and knowledgeable service staff, the hallmark of our company. Our technical support group is committed to handling all of your thermal analysis and rheology needs, and is available by phone, email and through the internet.
TA Instruments' reputation for high technology products, quality manufacturing and unbeatable after sales support is why more customers recommend TA products to their colleagues around the world. Headquartered in New Castle, DE, TA Instruments prides ourselves in the technical competence and professionalism that our sales force offers. We are the only thermal analysis, rheology, and microcalorimetry supplier recognized worldwide for our prompt, courteous and knowledgeable service staff, the hallmark of our company. Our technical support group is committed to handling all of your thermal analysis and rheology needs, and is available by phone, email and through the internet.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.