A DLS system’s performance depends on the sample. For example, while the upper size limit may be > 5 μm for an emulsion, it could be in the range of < 1 μm for a high density particle, owing to the lack of accuracy once sedimentation begins. The upper concentration limit may be > 40 vol.% for particles < 10 nm, but closer to 1% for particles ~ 1 μm.
Though the DLS system generates results for each analysis, there is no guarantee of equal accuracy in every result. The lower size and concentration limits for the Nicomp DLS systems, for instance, depend on both sample and configuration of the system. A higher power laser and high sensitivity APD detector is recommended in the case of low concentration measurements of smaller particles.
This article highlights the Nicomp DLS system’s ability to perform low concentration measurements of lysozyme.
Experimental Procedure
As depicted in Figure 1, lysozyme is a single chain polypeptide, consisting of 129 amino acids cross-linked with four disulfide bridges. With a DLS-measured particle size of roughly 3.6 nm, the molecular weight of this protein is 14,307 Da.
Figure 1. Lysozyme protein
As prior experience suggests, the monomer form of lysozyme displays high stability at low pH values close to pH 2.2. By first dissolving 14.91 g of KCl in 100 mL filtered DI water, a KCl buffer at pH 2.2 was prepared to create a 1.0.2 M KCl solution. This process was followed by the addition of 8.58 ml concentrated HCl into 491.4 ml water, in order to create 0.2 M HCl solution.
The next step involved adding 50 mL of the 0.2 M KCl solution into a 200 mL clean bottle, followed by the gradual addition of 7.8 mL of the 0.2 M HCL into the KCL solution. Finally, a total volume of 200 ml was obtained through the addition of filtered DI water. To filter this buffer solution for any of the subsequent steps, a 0.2 µm syringe filter was always utilized.
Purchased from Sigma Aldrich, the lysozyme used had a catalog number L6876, while lysozyme from a chicken egg was supplied as a lyophilized powder. Using UV absorbance, the protein content of the lysozyme used was 90%, with remaining constituents in the form of buffer salts, such as sodium chloride and sodium acetate.
Thus, all concentrations reported in this experiment are slightly less than by a factor of roughly 10%, considering that no concentration corrections were carried out. By adding 0.1 g lysozyme powder to 100 mL filtered buffer solution and then mixing it for 1 h, the base 10 mg/mL lysozyme sample was created.
Subsequently, this was then diluted at a ratio of 10:1 with filtered buffer solution to prepare 1 mg/mL concentration. As a result, the sample was then diluted at a ratio of 10:1 with filtered buffer to form the 0.1 mg/mL concentration sample.
The Nicomp DLS system is equipped to measure lysozyme down to a concentration of 0.1 mg/mL, i.e. the lower concentration limit for the instrument. As part of this experiment, an off the shelf Nicomp 380 DLS system was used, coupled with an APD detector and 658 nm, 50 mW laser.
All measurements stated were performed at 90º. Results are shown in Figures 2 and 3 – Figure 2 displays the system setup dialog box settings, while Figure 3 depicts the Particle Sizing Control Menu settings.
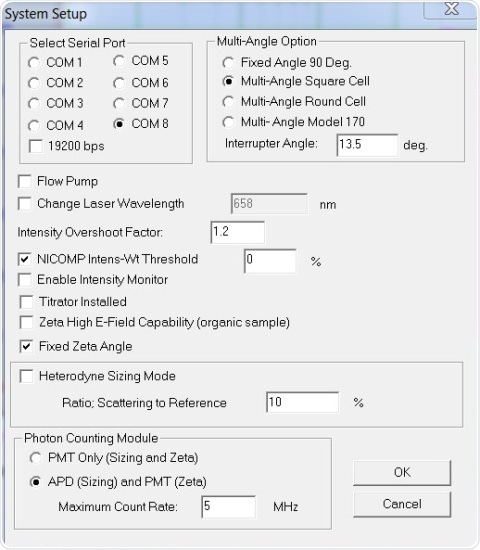
Figure 2. System Setup settings
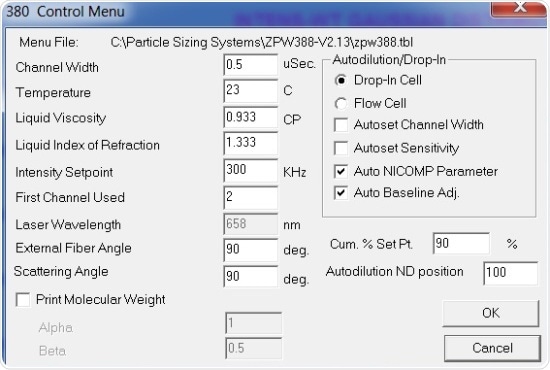
Figure 3. Control menu settings
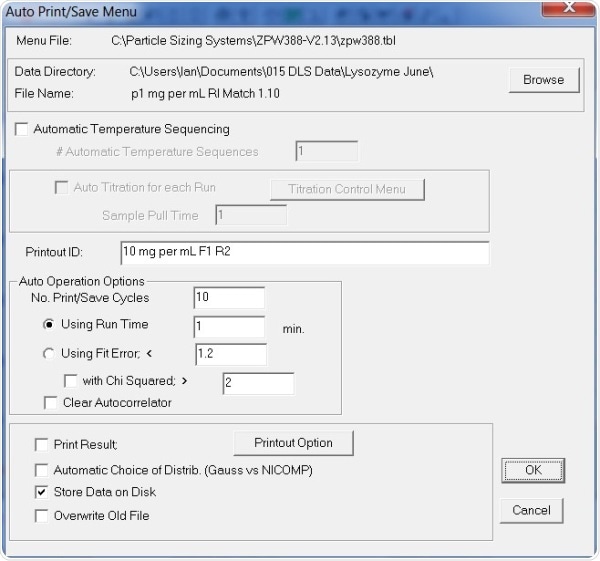
Figure 4. Auto Print/ Save Menu settings
Experimental Results
Through a 0.2 µm syringe filter, the 10 mg/mL lysozyme sample was filtered and then introduced into a clean square glass cuvette. Using the settings mentioned above, the sample analysis was performed, with Figure 5 depicting the experimental results acquired.
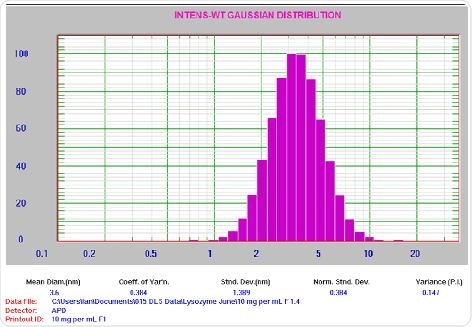
Figure 5. 10 mg/mL lysozyme result. For clarification: Mean Diam = 3.6 nm, Std Dev = 1.389 nm, PI = 0.147.
It is important to note that the PI value is small as anticipated for a well- prepared sample with count rate ~160 kcps. Further, the 1 mg/mL lysozyme sample was filtered through a 0.2 µm syringe filter and subsequently introduced into a clean square glass cuvette. Following this, aggregate separation was conducted by centrifuging the cuvette at 8000 rpm for 10 min. Finally, the sample analysis was performed using the aforementioned settings. Figure 6 displays the corresponding results.
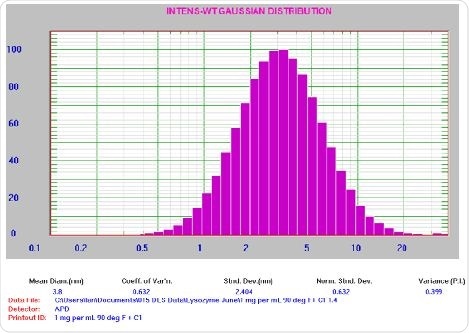
Figure 6. 1.0 mg/mL lysozyme result. For clarification: Mean Diam = 3.8 nm, Std Dev = 2.404 nm, PI = 0.399.
The presence of possible aggregates is represented by the fact that the PI value is higher than anticipated for a well-prepared sample. Such a scenario is typical for a sample at low count rate (~30 kcps). Following this, the 0.1 mg/mL lysozyme sample was filtered through the 0.2 µm syringe filter and introduced into a clean round glass cuvette. After this, centrifuging of the cuvette at 8000 rpm was conducted for 10 min for aggregate separation.
The next step involved placing the cuvette in a square cell consisting of oil with matching refractive index (RI). This was done to enhance the reproducibility of the lowest concentration measurements. Finally, using the instrument settings, the sample analysis was performed. Figure 7 reports the corresponding results.
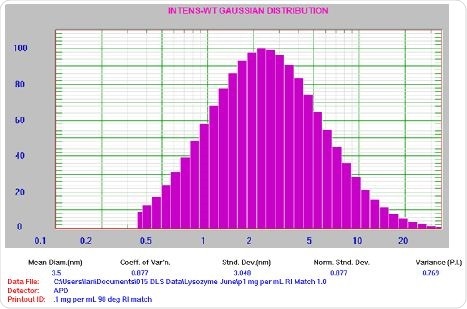
Figure 7. 0.1 mg/mL lysozyme results. For clarification: Mean Diam = 3.5 nm, Std Dev = 3.048 nm, PI = 0.769.
An important point to be noted is that the PI value is greater than predicted for a well-prepared sample. This implies the presence of possible aggregates, a scenario that is typical for a sample at low count rate (~19 kcps).
As seen in the results displayed above, there is a definite advantage of using the RI matching cell to enhance the count rate for the lowest concentration experiments. Figure 8 depicts the repeatability of the instrument, which was assessed by measuring the 0.1 mg/mL sample repeatedly.
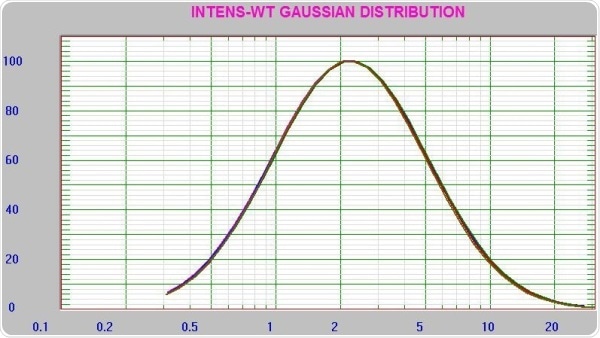
Figure 8. Nine consecutive results at 0.1 mg/mL concentration lysozyme.
Conclusion
This article presented all results presented in intensity distribution by applying the Gaussian distribution results. In other words, no other changes or conversions were utilized to obtain these results. Thus, as is evident from the results, it is clear that the Nicomp DLS system can measure lysozyme at the specified concentration of 0.1 mg/mL.
About Entegris
Since our inception in 1978 we have been providing leading edge instrumentation for the field of particle size analysis. We are an applications driven company that provides solutions to our customer's most complex particle sizing and zeta potential monitoring problems. From wet to dry, online to research laboratory we have engineered a complete family of modular designed instruments that can be configured to meet the specific demands of an application.
Single Particle Optical Sizing (SPOS)
The AccuSizer operates using the principle of single particle optical sizing (SPOS). This technique generates high accuracy, high resolution particle size distribution results as well as concentration data in particles/mL. The AccuSizer platform includes multiple configurations designed to meet a wide array of applications including laboratory and online systems. Multiple sensor options cover a wide range of particle size and concentration limits. Advanced fluidics modules can handle low concentration samples requiring no dilution to highly concentrated inks and CMP slurries using our patented two stage exponential dilution system.
Dynamic Light Scattering
The Nicomp dynamic light scattering (DLS) system measures particle size below 1 nm and zeta potential to study particle charge. The unique Nicomp algorithm provides the highest resolution results for samples containing more than one peak. Other unique capabilities include auto-dilution and auto-samplers to facilitate high sample throughput.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.