Of all the organs, the liver is most susceptible to drug toxicity. Drug-induced liver injury (DILI) is a leading cause of drug attrition, with over 750 FDA-approved drugs known to have a level of DILI risk.1
Numerous strategies to de-risk DILI in drug discovery have been developed and, recently, the focus has shifted to more reliable human in vitro 3D liver models that can better predict DILI.2 These models use primary human liver cells that are cultured together in a physiologically relevant environment, enabling them to remain functional for extended periods.3
In this study, CN Bio assessed whether an in vitro Liver-on-a-Chip, also known as a human liver microphysiological system (MPS), could be utilized as an accurate predictor of DILI risk over a wide set of compounds. Thirteen of these compounds are known as mildly and severely hepatotoxic pharmaceutical drugs. The study also included two non-approved antisense oligonucleotides (ASOs).
The liver MPS is utilized in CN Bio’s PhysioMimix DILI assay. It is made up of primary human hepatocytes (PHH) and Kupffer cells (HKC), cultured under flow perfusion on 3D scaffolds and can maintain highly functional 3D liver microtissues for up to four weeks, should longer-term repeat dosing studies be required. The potential of CN Bio’s PhysioMimix DILI assay to accurately identify drugs with varying risk has been demonstrated in three case studies.
The first study analyzes two pharmaceutical drugs, Tolcapone and Entacapone, which are typically used for Parkinson’s disease management. Both are classified as catechol-O-methyltransferase (COMT) inhibitors. Tolcapone, the first FDA-approved COMT inhibitor, was introduced in 1998 to the US market. Unfortunately, its DILI risk was missed during preclinical safety testing. Severe liver complications were observed in the clinic that caused the death of some patients and the drug's withdrawal soon after. In 2009, tolcapone was re-introduced to the market but only as a last resort for Parkinson’s disease treatment.5
Conversely, the structural analog to Tolcapone, Entacapone, has a low hepatoxicity risk and was the second COMT inhibitor to be approved for Parkinson’s disease. It is deemed relatively safe but has been known to cause liver damage occasionally.6
Published data from in vitro cellular assays demonstrated that both compounds induce mitochondrial dysfunction in a dose-dependent manner or through the inhibition of efflux transporters.7 Tolcapone was associated with immune-mediated DILI (iDILI), which caused a sharp increase in T-regulatory cells.8, whereas Entacapone did not.
The second case study is focused on two antidiabetic thiazolidinediones: Troglitazone and Pioglitazone. Classified as having severe DILI concern, Troglitazone was licensed in 1997 to treat type 2 diabetes. The FDA withdrew the approval just three years later, due to repeated reports of liver injury, including acute liver failure, in the post-marketing surveillance.
Published animal studies failed to predict Troglitazone’s ability to cause severe liver injury. This compound’s toxicity was also undetected in traditional in vitro 2D hepatic assays.9
Pioglitazone is a compound known to be of low-DILI concern. Examples of acute liver injury due to pioglitazone are uncommon but have been reported occasionally during post-marketing in patients with heart failure or obesity.10,11 During development, pioglitazone hepatotoxicity was not observed in traditional in vitro 2D primary hepatocyte cultures or more advanced 3D models.12,13
The last case study examines the capability of CN Bio’s PhysioMimix DILI assay to predict the DILI risk of two non-approved ASOs: LNA32 and LNA43. Both were shown to be safe or very hepatotoxic using in vivo rodent models respectively.14,15
ASOs are engineered molecules that contain locked nucleic acid (LNA) made to instigate target RNA cleavage. Numerous studies show a strong link between liver toxicity and the sequence content of the LNA gapmer of ASOs.16 The liver serves as an accumulation target organ where ASO-mediated toxicity is likely observed.14
In vitro biotransformation studies, performed at an early stage of drug development, are essential for predicting the potential hepatotoxicity of ASOs. For modalities like ASOs, where there is a human-specific mode of action, animal models are less suitable. To eliminate false reporting due to interspecies differences, it is paramount to make use of the most human-translatable in vitro models available during preclinical evaluation.
In this study, CN Bio’s PhysioMimix and Multi-chip Liver-12 plates were used to culture Liver MPSs. Following drug dosing, a variety of functional liver-specific endpoints were analyzed (including clinical biomarkers such as serum albumin and alanine aminotransferase (ALT) across a dose-response to generate EC:50 curves. Analysis of these functional liver-specific endpoints enabled the creation of distinct mechanistic “signatures of hepatotoxicity” to show the assay’s ability to assess the human DILI risk of differing drug modalities.
Aims
- Demonstrate the ability of The PhysioMimix DILI assay to produce clinically relevant, high-content data (six endpoint EC:50 curves) from a wide set of thirteen compounds from mildly to severely hepatotoxic.
- Demonstrate the ability to accurately evaluate the DILI risk of novel modalities through the generation of clinically relevant, high-content data (six endpoint EC:50 curves) for two ASOs.
- Conclude whether the assay can more accurately predict human DILI over conventional preclinical approaches.
Results

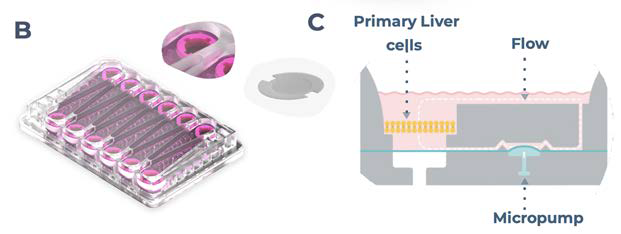
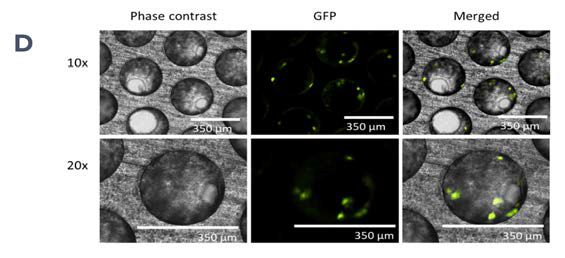
Figure 1. PhysioMimix enables the generation of 3D human liver microtissues, functional for up to four weeks. A) In vitro liver microtissues are generated by the PhysioMimix, which uses open well plates designed for the 3D culture of primary liver cells on an engineered scaffold. B) Schematic representation of a PhysioMimix Multi-chip Liver-12 plate for the coculture of PHHs and HKCs. C) Cross-section of a well indicating the scaffold and fluidic flow perfusion of 3D liver microtissues by micropumps. D) Phase contrast microscopy (10 x and 20 x) and immunofluorescence (IF) labeling of 3D liver MPS. To visualize the HKCs, before seeding HKCs were transduced with an adenoviral vector expressing eGFP. Representative photomicrographs are shown. The transduction and imaging were performed as a standalone experiment to demonstrate cell localization. HKCs cells are pre-validated in-house before use in experimental cell culture, and must have low levels of post-thaw activation; this is assessed by measuring biomarkers IL-6 and TNF-α. Image Credit: CN Bio
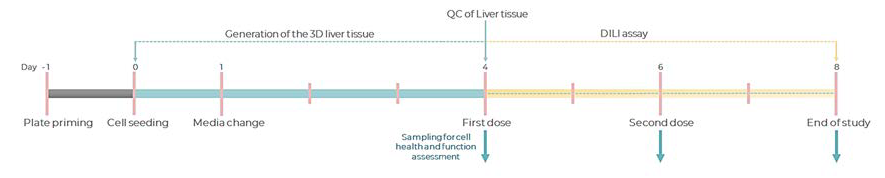
Figure 2.Experimental timeline of a standard DILI assay in the PhysioMimix Multi-chip Liver-12 plate. Image Credit: CN Bio
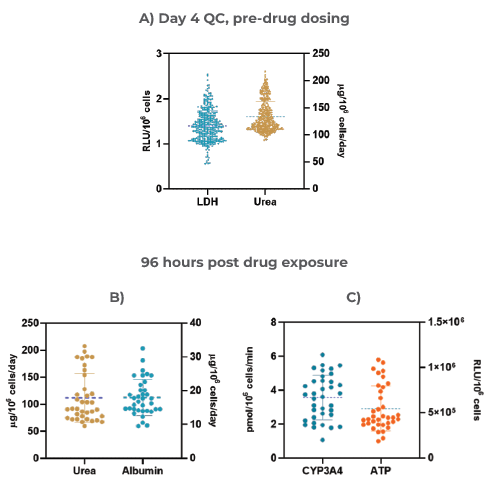
Figure 3. The Liver MPS produces highly reproducible data and consistent microtissues A) 3D Liver microtissue quality control (QC) metrics at Day four (Mean ± standard deviation (SD), N = 360); Functionality assessment at the end of the drug dosing (96 hours) – B) Albumin and Urea, and C) CYP3A4 and ATP. Data was collected from 12 individual experiments; for each experiment, three vehicle control replicates were used (data shown are Mean ± SD, N = 36).Image Credit: CN Bio
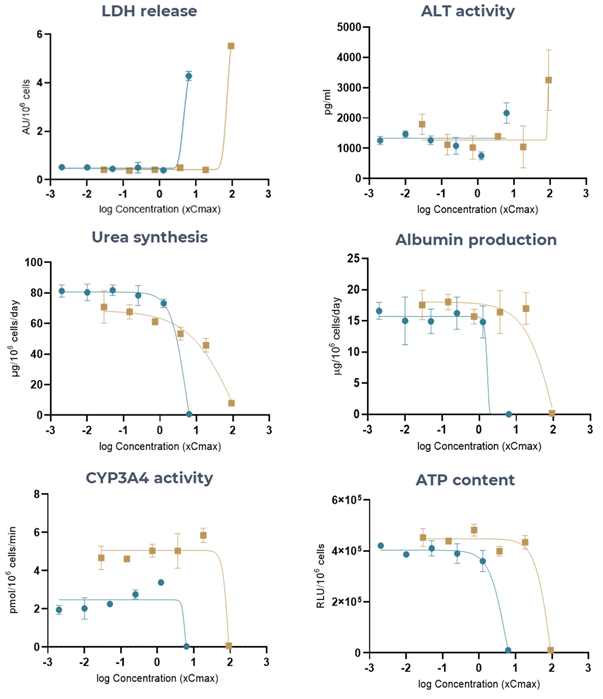
Figure 4. The Liver MPS generates accurate Tolcapone (high-DILI-concern) and Entacapone (low-DILI-concern) DILI profiles, using multiple hepatotoxic endpoints. Liver microtissues were exposed to Tolcapone (blue) and Entacapone (brown) for 96 hours. All endpoint measurements were derived from the same liver MPS. Data shown are mean ± SD, N = 3, and all from 96-hour samples, apart from LDH release which was measured at 48 hours. Image Credit: CN Bio
Table 1. Toxicity of Tolcapone and Entacapone in 3D human liver MPS microtissues. Liver microtissues in the MPS were exposed to both compounds for 96 hours. LDH data was measured at 48 hours post-dose. Data were generated from seven-point dose responses, with N = 3 per concentration. Maximum test concentration for Tolcapone = 6.3 x Cmax and for Entacapone = 91.5 x Cmax. Source: CN Bio
| Assay |
Tolcapone |
Entacapone |
| |
EC50 (xCmax) |
R2 |
EC50 (xCmax) |
R2 |
| LDH |
4.9 |
0.9 |
73.8 |
0.9 |
| ALT |
3.3 |
0.6 |
87.1 |
0.7 |
| Albumin |
0.3 |
0.9 |
62.5 |
0.9 |
| Urea |
4.5 |
0.9 |
39.3 |
0.9 |
| CYP3A4 |
5.5 |
0.7 |
82.5 |
0.9 |
| ATP |
7.2 |
0.9 |
75.7 |
0.9 |
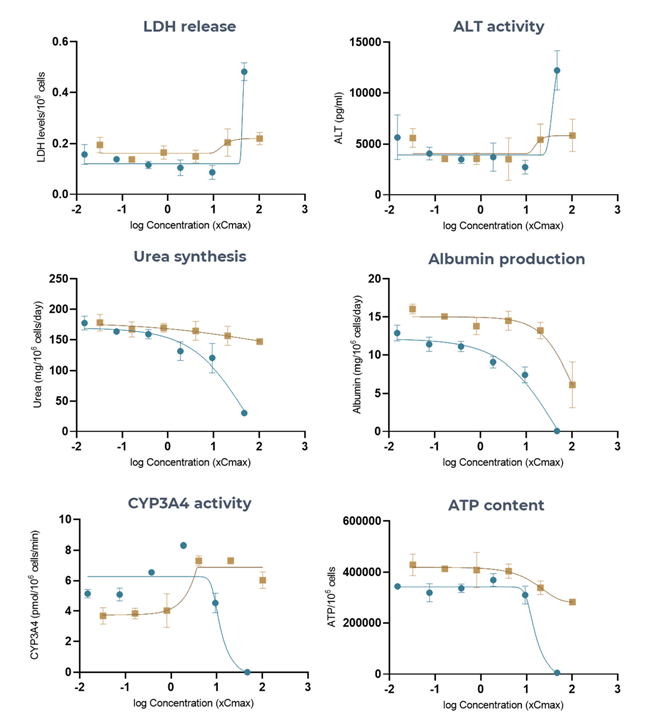
Figure 5. The Liver MPS accurately generates troglitazone (high-DILI-concern) and pioglitazone (low-DILI-concern), DILI profiles using multiple hepatotoxic endpoints. Liver microtissues in the MPS were exposed to troglitazone (blue) and pioglitazone (brown) for 96 hours. All endpoint measurements were derived from the same liver MPS culture. Data shown are mean ± SD, N = 3, and all from 96-hour samples. Image Credit: CN Bio
Table 2. Toxicity of Troglitazone and Pioglitazone in 3D human liver MPS microtissues. Liver microtissues in the MPS were exposed to both compounds for 96 hours. Data were generated from seven-point dose responses, with N = 3 per concentration. Maximum test concentration for troglitazone = 47 x CCmax, maximum test concentration for pioglitazone = 100 xCmax. Source: CN Bio
|
| Assay |
Tolcapone |
Entacapone |
| |
EC50 (xCmax) |
R2 |
EC50 (xCmax) |
R2 |
| LDH |
43.9 |
0.9 |
ND |
| ALT |
40.3 |
0.8 |
ND |
| Albumin |
21 |
0.9 |
63.2 |
0.7 |
| Urea |
22.7 |
0.9 |
ND |
| CYP3A4 |
11.8 |
0.8 |
ND |
| ATP |
15.5 |
0.9 |
16.5 |
0.7 |
ND - not detected
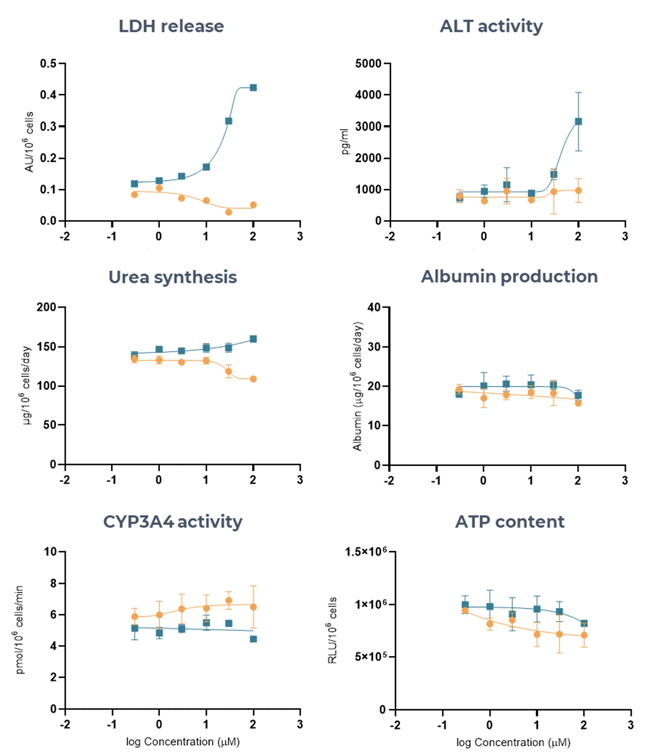
Figure 6. Liver MPS accurately determines the DILI risk of two ASOs (LNA43, LNA32), using multiple hepatotoxic endpoints. Liver microtissues in the MPS were exposed for 96 hours to LNA43 (blue), known to be severely hepatotoxic in vivo, and LNA32 (brown), known to be safe in vivo. All endpoint measurements were derived from the same liver MPS culture. Data shown are mean ± SD, N = 3, and all from 96-hour samples. Image Credit: CN Bio
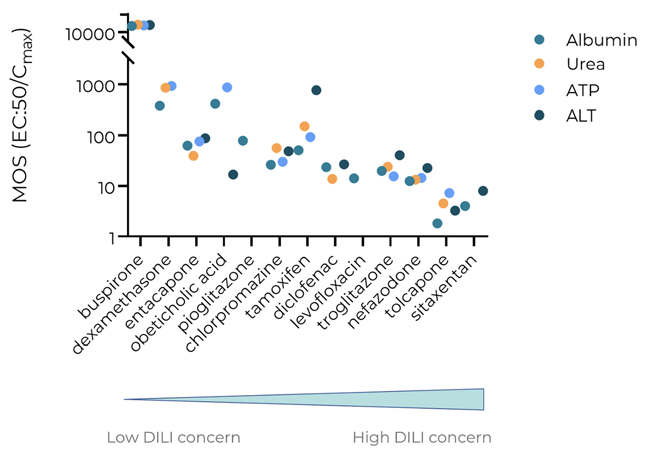
Figure 7. The PhysioMimix DILI assay accurately determines the liability of a broad set of known severely and mildly hepatotoxic compounds. Exposure-corrected cytotoxicity or margin of safety (MOS = EC:50/Cmax) was determined for four key biomarkers albumin, urea, ATP, and clinical biomarker ALT, following 96 hours of exposure. All endpoint measurements were derived from the same liver MPS culture. Data shown are mean ± SD, N = 3. Tested compounds are arranged based on DILI rank, from low DILI concern (left) to high DILI concern (right). Image Credit: CN Bio
Conclusion
The PhysioMimix assay was able to accurately predict human DILI for all compounds tested. With its heightened sensitivity, the model identified hepatotoxicants (Pioglitazone, Dexamethasone, Levofloxacin) that were missed by 2D and some 3D in vitro models.5 The experimental approaches and data show that CN Bio’s PhysioMimix cultures metabolically active and highly functional liver microtissues.
These liver microtissues can be consistently and reliably employed to predict, with accuracy, the DILI liability of drugs of differing modalities. CN Bio hypothesizes that by recapitulating the immune aspects of the liver that cause DILI, the assay's sensitivity was improved.
The model can be additionally modified to include adaptive immune cells, like Peripheral Blood Mononuclear cells (PBMCs), into the recirculating flow that perfuses the MPS model to provide an immunocompetent solution.
Through the measurement of six different endpoint biomarkers, including clinical biomarkers (ALT and serum albumin), the model produced a “signature of hepatotoxicity” for each test article to assist with identifying compounds with various levels of DILI concern, and reveal mechanisms of their toxicity.
The human translatable data generated by the PhysioMimix DILI assays enables researchers to progress only the safest drugs into in vivo studies, minimizing the number of animals required. The approach provides a viable alternative to animal models where translatability to humans is predicted to be poor and for the testing of advanced drug modalities where human-specific targets and pathways are required to be better prepared for the clinic.
References and further reading
- Dirven, H. et al. (2021). Performance of preclinical models in predicting drug-induced liver injury in humans: a systematic review. Sci. Rep. 11, 1–19.
- Walker, P. A., Ryder, S., Lavado, A., Dilworth, · Clive & Riley, R. J. (2020). The evolution of strategies to minimise the risk of human drug-induced liver injury (DILI) in drug discovery and development. Arch. Toxicol. 94, 2559–2585.
- Rubiano, A. et al. (2021). Characterizing the reproducibility in using a liver microphysiological system for assaying drug toxicity, metabolism, and accumulation. Clin Transl Sci 14.
- Rowe, C. et al. (2018). Perfused human hepatocyte microtissues identify reactive metabolite-forming and mitochondria-perturbing hepatotoxins. Toxicol. Vitr. 46, 29–38.
- Artusi, C. A., Sarro, L., Imbalzano, G., Fabbri, M. & Lopiano, L. Safety and efficacy of tolcapone in Parkinson’s disease: systematic review. doi:10.1007/s00228-020- 03081-x/Published.
- Kuoppamäki, M., Leinonen, M. & Poewe, • Werner. Efficacy and safety of entacapone in levodopa/carbidopa versus levodopa/benserazide treated Parkinson’s disease patients with wearing-off. J. Neural Transm. 122,.
- Longo, D. M., Yang, Y., Watkins, P. B., Howell, B. A. & Siler, S. Q. (2016). Elucidating differences in the hepatotoxic potential of tolcapone and entacapone with DILIsym®, a mechanistic model of drug-induced liver injury. CPT Pharmacometrics Syst. Pharmacol. 5, 31–39.
- Teschke, R., Chen, M., Kenna, J. G. & Uetrecht, J. (2019). Article 837 REVIEW Frontiers in Pharmacology | www.frontiersin.org Citation: Uetrecht J (2019) Mechanistic Studies of Idiosyncratic DILI: Clinical Implications. Front. Pharmacol 10, 837.
- Bell, C. C. et al. (2018). Comparison of Hepatic 2D Sandwich Cultures and 3D Spheroids for Long-term Toxicity Applications: A Multicenter Study. Toxicol. Sci. 162, 655–666.
- Chen, M., Suzuki, A., Borlak, J., Andrade, R. J. & Lucena, M. I. (2021). Drug-induced liver injury: Interactions between drug properties and host factors. J. Hepatol. 63, 503–514 (2015).
- Richardson, B., Khan, M. Q., Brown, S. A., Watt, K. D. & Izzy, M. Personalizing Diabetes Management in Liver Transplant Recipients: The New Era for Optimizing Risk Management. Rev. | Hepatol. Commun. 6, 2022.
- Li, F., Cao, L., Parikh, S. & Zuo, R. (2020). Three-Dimensional Spheroids With Primary Human Liver Cells and Differential Roles of Kupffer Cells in Drug-Induced Liver Injury. J. Pharm. Sci. 109, 1912–1923.
- Proctor, W. R. et al. (2017). Utility of spherical human liver microtissues for prediction of clinical drug-induced liver injury. Arch. Toxicol. 91, 2849–2863.
- Dieckmann, A. et al. (2018). A Sensitive In Vitro Approach to Assess the HybridizationDependent Toxic Potential of High Affinity Gapmer Oligonucleotides. Mol. Ther. - Nucleic Acids 10, 45–54.
- Sewing, S. et al. (2016). Establishment of a predictive in vitro assay for assessment of the hepatotoxic potential of oligonucleotide drugs. PLoS One 11, 1–15.
- Burel, S. A. et al. (2015). Hepatotoxicity of high affinity gapmer antisense oligonucleotides is mediated by RNase H1 dependent promiscuous reduction of very long pre-mRNA transcripts. Nucleic Acids Res. 44, 2093–2109.
- Bell, C. C. et al. Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease OPEN. Nat. Publ. Gr. (2016) doi:10.1038/srep25187.
About CN Bio
CN Bio is a leading organ-on-a-chip (OOC) company that offers a portfolio of products and contract research services to optimize the accuracy and efficiency of bringing new medicines to market. With more than a decade of research and development experience, they aim to transform the way human-relevant pre-clinical data is generated through the development of advanced in vitro human organ models.
CN-Bio's PhysioMimix® Core microphysiological system (MPS) enables researchers to recreate human biology in the lab and is the only microphysiological system with validated performance across single-, multi-organ, and higher throughput configurations. This easy to adopt, adapt and scale technology bridges the gap between traditional cell culture and human studies, to support the development of safer and more efficacious therapeutics, whilst reducing the dependence on animal model usage.
CN Bio’s portfolio of products (MPS, 3D validated cells, consumable plates) and services support researchers that require reliable, data-rich, in vitro studies, to uncover novel mechanistic insights into drug or disease mechanism of action.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.