The accessory protein open-reading frame-6 (ORF6) expressed by the causal virus of the coronavirus disease 2019 (COVID-19) pandemic, SARS-CoV-2, has been indicated by various studies to play a vital role in viral pathogenicity and viral modulation of host cells. However, there is insufficient data about the function of ORF6 and its impact on the host cell environment.
Study: SARS-CoV-2 ORF6 disrupts innate immune signalling by inhibiting cellular mRNA export. Image Credit: Design_Cells / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
About the study
The present study examined the impact of the SARS-CoV-2 accessory protein ORF6 on the human cell environment to promote SARS-CoV-2 replication and inhibit antiviral responses.
Inactivated fetal bovine serum (FBS) and penicillin or streptomycin were added to Vero cell and Mus dunni tail fibroblast (MDTF) lines. These stock cells underwent testing to detect mycoplasma contamination and were verified by short-tandem repeat (STR) profiling. Two distinct plasmids were employed to express individual SARS-CoV-2 ORF proteins. Vector encoding of individual ORF proteins was performed, and ORF genes were synthesized, followed by cloning of the genes into plasmids. ORF6 mutations were then introduced into the plasmids, and murine leukemia virus (MLV) and human immunodeficiency virus 1 (HIV-1) virus-like particles (VLPs) were produced by co-transfecting the cell lines with plasmids.
Immunoblotting was performed by lysing the cells in radioimmunoprecipitation assay (RIPA) buffer containing deoxyribonuclease (DNase) and protease inhibitor cocktail. The protein lysates were boiled, separated, and transferred to a polyvinylidene fluoride (PVDF) membrane. Primary and secondary antibodies were used to analyze the lysed proteins by immunoblotting.
Protein content in the lysed cells was evaluated by bicinchoninic acid (BCA) assay. The RNA extracted from the total cell lysates was processed with DNase. A portion of the RNA was then converted to complementary DNA (cDNA) and tested with quantitative polymerase chain reaction (qPCR) analysis.
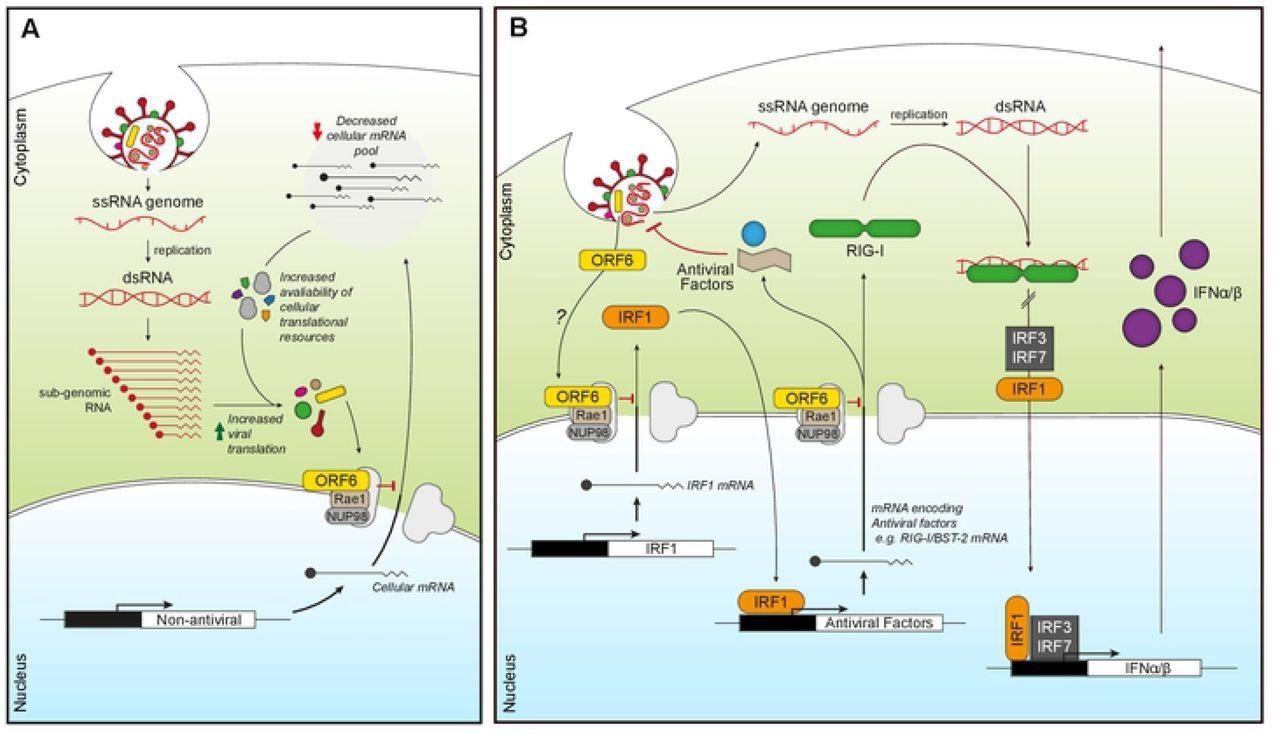
ORF6 inhibits mRNA export to favour viral translation and suppress innate signaling. (A) Incoming or expressed ORF6 (yellow rectangles) binds to the Nup98-Rae1 complex (grey/brown) blocking the export of cellular mRNA (black lines). This subsequently reduces the cellular mRNA pool, increasing the availability of cellular translational machinery for viral translation as well as decreasing the expression of cellular proteins. (B) Incoming or expressed ORF6 (yellow) binds to the Nup98-Rae1 complex (grey/brown), inhibiting export of cellular mRNA encoding IRF1 (orange). This prevents the translation of IRF1, blocking IRF1 regulation of the transcription of additional steady-state antiviral factors, like RIG-I and BST-2. ORF6 also inhibits nuclear export of mRNA encoding RIG-I (green), preventing detection of viral dsRNA produced during coronavirus replication. This helps reduce IRF1/3/7 activity and subsequent transcription of IFNα/β (purple), which prevents IFNα/β inducing an antiviral state in an autocrine and paracrine manner.
Results
The study results showed that all VLPs had a minimum of 104 infectious units/ml, except for the ORF6-encoding VLP, which led to the inhibition of the production of HIV-1 structural polyprotein, Gag. This decrease in Gag expression was proportional to the increase in doses of ORF6. A similar trend was observed in the reduction of HIV-1 VLP titers with increasing amounts of ORF6 compared to the control sample.
The research suggested that ORF6 interacted with the cellular protein Rae1, an mRNA export factor, and inhibited protein expression. Also, the interaction of ORF6 with the Rae1-Nup98 complex was dependent on Rae1, and there was no direct interaction between ORF6 and Nup98. It was also noted that the impact of ORF6 on the expression of proteins was mainly affected by Rae1, indicating the inhibition of mRNA export.
ORF6 reduced the nuclear export of green fluorescent protein (GFP) mRNA depending on the amount of the Rae1 gene present. In the absence and presence of interferon (IFN) signaling, 2,431 and 2,128 nucleus-enriched mRNA species, respectively, were observed while 180 and 81 were cytoplasm-enriched with ORF6, respectively. This indicated that ORF6 inhibited the export of exogenous mRNA as well as the nuclear export of a wide range of cellular mRNA.
A total of 134 mRNA species were upregulated after IFN treatment in either the cytoplasm and/or in the nucleus and were hence called interferon upregulated genes (IUGs). These IUGs encoded proteins that regulated the immune response or inhibited viral replication. The presence of ORF6 caused the downregulation of 69 IUGs in the cytoplasm and the upregulation of two IUGs, indicating that ORF6 can prevent mRNA expression of IUGs.
Research showed that ORF6 could inhibit mRNA export in SARS-CoV-2 infections. Also, the mRNA retention caused by SARS-CoV-2 was similar to the retention affected by ORF6 alone, which inhibited the export of mRNA species that caused transcription and immune regulation. This suggested the ability of ORF6 to impact cellular pathways involved in viral infections.
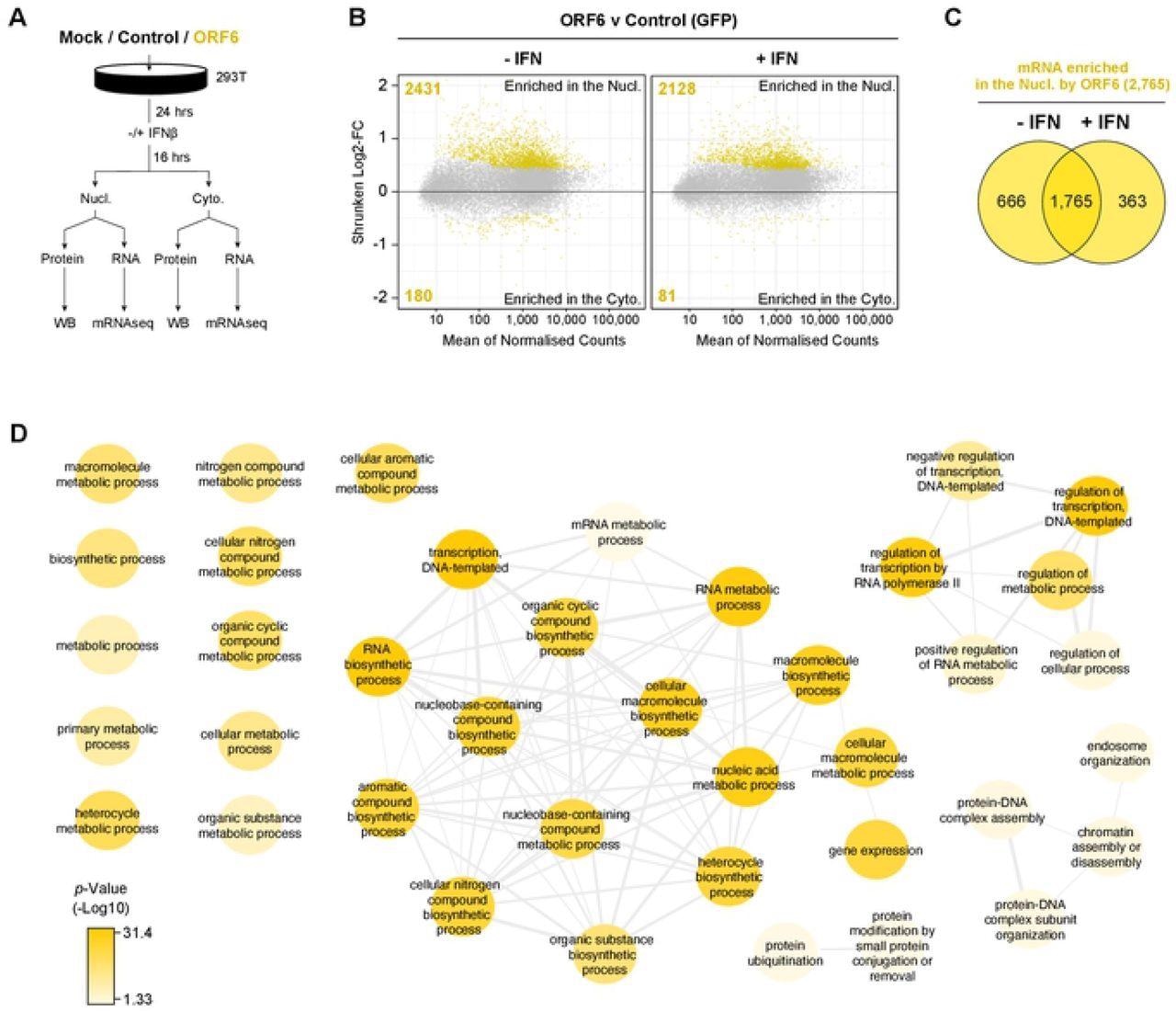
ORF6 inhibits the nuclear export of cellular mRNA. HEK293T cells were transfected with pLVX-EF1α-GFP or pLVX-EF1α-ORF6 or left untransfected (mock). After 24h, cells were either treated with IFN-β (1,000 units/ml) for 16 hrs or left untreated. Cells were then harvested and fractionated into nuclear (Nucl) and cytoplasmic (Cyto) fractions. Cells were either lysed for immunoblot analysis (see S4A Fig) or RNA was extracted for mRNAseq. (A) Schematic of experiment design. (B) The log2-fold change (Log2-FC) in mRNA abundance was calculated between the Nucl and Cyto fractions for both GFP and ORF6 expressing cells, without and with IFN treatment (see S4B Fig). The log2-FC in mRNA abundance was then directly compared between ORF6 and GFP, without (left panel) and with (right panel) IFN, to determine which mRNAs were specifically enriched in the nucleus or cytoplasm by ORF6. The log-2FC was weighted against the adjusted p-value (shrunken Log2-FC) to show significantly enriched mRNAs (yellow points). The number of mRNAs significantly enriched is shown. The mRNA count was normalized and averaged between the three biological repeats. (C) Venn diagram of the number of mRNAs that were significantly enriched in the nucleus by ORF6 with and without IFN. (D) mRNAs significantly enriched by ORF6 without IFN treatment were analyzed for enriched GO terms, which were filtered and mapped using the REVIGO tool. Highly similar GO terms are grouped and linked. Colour intensity reflects significance levels.
Conclusion
The study findings concluded that ORF6 could significantly inhibit protein production and block mRNA export into the cytoplasm from the nucleus, which led to effective blockage of protein translation. In addition, ORF6 also inhibited the export of a broad spectrum of viral RNAs, including antiviral factor-encoding genes, indicating the disruption of host cell immune signals due to SARS-CoV-2 ORF6.
Furthermore, this inhibition of mRNA export suggests that the accessory protein may inhibit immune signaling in the early stages of antiviral response by repressing both antiviral transcription factors and pathogen recognition receptors (PRRs) expression. The researchers believe that understanding the mechanism of immune response inhibition of viral ORF6 will facilitate the development of new therapeutic strategies against SARS-CoV-2 infections.
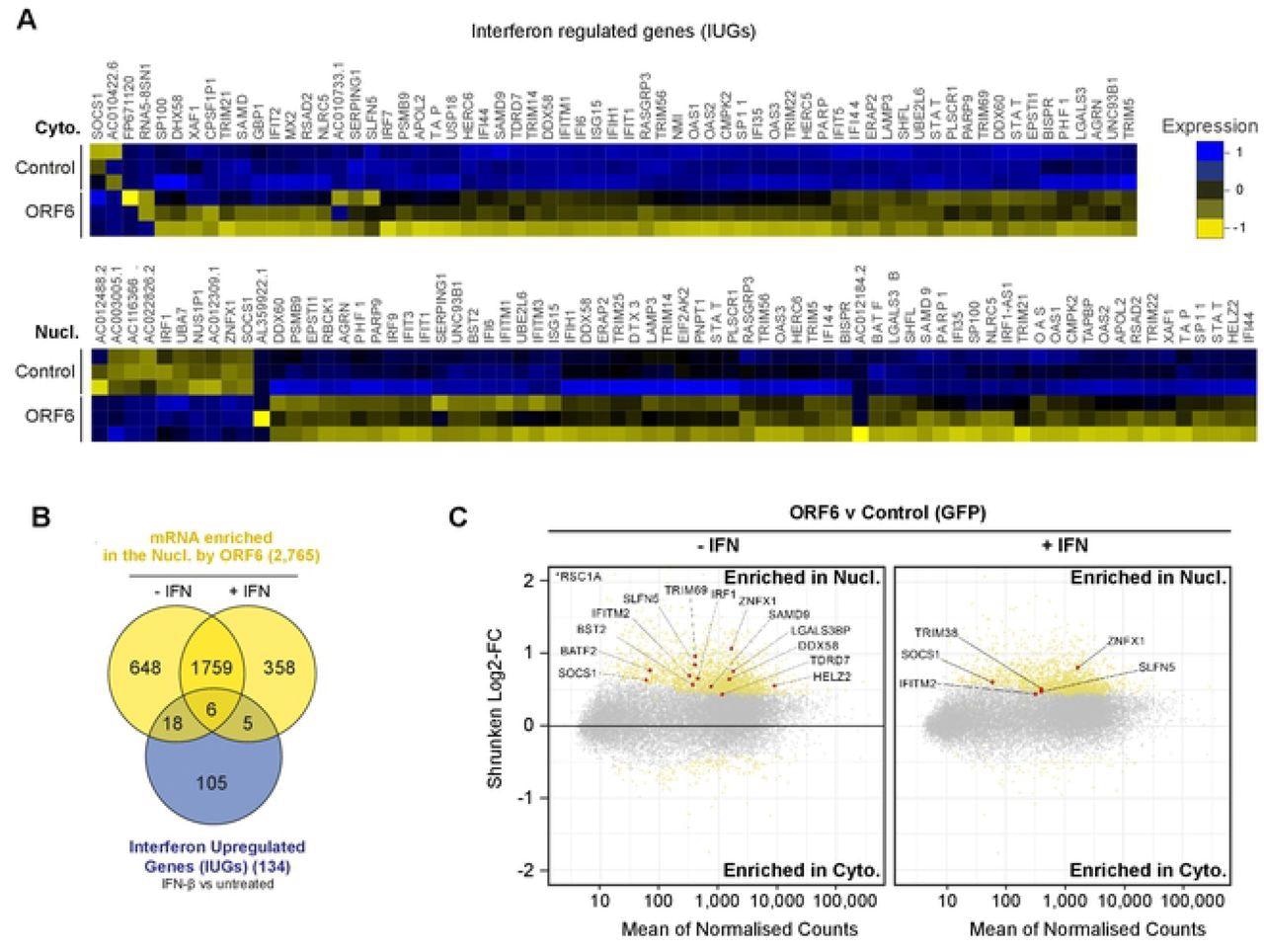
ORF6 inhibits the nuclear export of IFN-upregulated mRNA. Interferon upregulated genes (IUGs) were identified as mRNAs that were significantly upregulated after IFN treatment compared to untreated cells (see S5A Fig). These IUGs were then compared to the list of mRNAs that were significantly upregulated or downregulated by ORF6 compared to the GFP control (see S5E Fig), in either the Cyto (upper panel) or Nucl (lower panel) fractions. The heat maps show the relative Log2-Fold Change in IUG expression in cells expressing GFP (Control) or ORF6 after IFN treatment, for three biological repeats. Venn diagram showing the number of IUG mRNAs that were significantly enriched in the nucleus by ORF6 with and without IFN treatment. (C) The log2-fold change (Log2-FC) in mRNA abundance was calculated between the Nucl and Cyto fractions for both GFP and ORF6 expressing cells, without and with IFN treatment, as in Fig 3B, to determine which mRNA were specifically enriched by ORF6. The log-2FC was weighted against the adjusted p-value (shrunken Log2-FC) to show significantly enriched mRNA (yellow points). IUGs are highlighted in red and labelled.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
SARS-CoV-2 ORF6 disrupts innate immune signaling by inhibiting cellular mRNA export, Ross Hall, Anabel Guedán, Melvyn W. Yap, George R. Young, Ruth Harvey, Jonathan P. Stoye, Kate N. Bishop, bioRxiv 2022.02.08.479664, doi: https://doi.org/10.1101/2022.02.08.479664, https://www.biorxiv.org/content/10.1101/2022.02.08.479664v1
- Peer reviewed and published scientific report.
Hall, Ross, Anabel Guedán, Melvyn W. Yap, George R. Young, Ruth Harvey, Jonathan P. Stoye, and Kate N. Bishop. 2022. “SARS-CoV-2 ORF6 Disrupts Innate Immune Signalling by Inhibiting Cellular MRNA Export.” Edited by Bart L. Haagmans. PLOS Pathogens 18 (8): e1010349. https://doi.org/10.1371/journal.ppat.1010349. https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1010349.