This article and associated images are based on a poster originally authored by Adam Buckle, Julia Unsicker and Iain McWilliam and presented at ELRIG Drug Discovery 2025 in affiliation with Arrayjet Ltd.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

The Arrayjet SMM-10 is a miniaturized small molecule library-on-a-chip for rapid drug discovery against complex drug targets, including structured RNA. Small molecule microarray (SMM) is an established hit discovery tool, which Arrayjet is expanding access to using its Mercury printing technology.
SMM-10 chips are offered ready-to-use, allowing a first exploration of Arrayjet’s 350K+ machine-learning filtered SM libraries.
This poster describes how the 10,000 compounds on the chip were selected for drug-like properties, with 5,000 chosen compounds for broad chemical diversity and 5,000 compounds selected for RNA-binding characteristics. We further demonstrate the flexibility of the SMM-10 screening methodology and successfully identify small molecule hits against both published and novel structured RNA drug targets.
Process overview: Small molecule microarrays
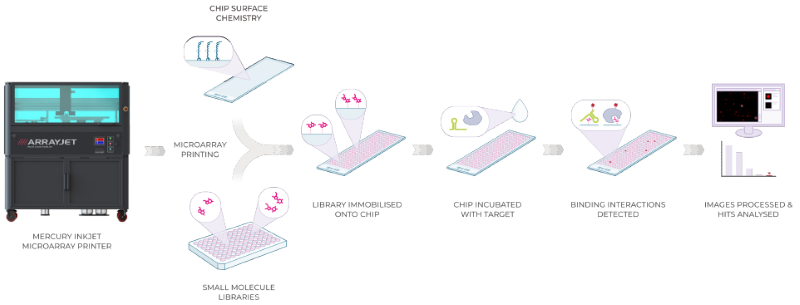
Figure 1. An overview of the Small Molecule Microarray (SMM) process. SMM follows a biochemical binding assay protocol. First, the glass chip surface is functionalized, typically with an isocyanate coating, to be highly reactive and allow the immobilization of a diverse range of SMs from a selected compound library. SMM uses unmodified compounds directly from standard library formats and concentrations, typically 10 mM in 100 % DMSO. Using Arrayjet's Mercury microarrayers to carry out microarray printing, each SM in the library is covalently coupled to the chip surface as an individual spot in a spatially resolved microscopic 2D grid. The SMM chip is then incubated with a target of interest, thereby screening for binding against the whole library in parallel. Binding interactions are identified through the detection of the target molecule fluorescence signal at a discrete spatial position on the microscopic grid. SMM is compatible with various target types, including proteins, structural RNA, DNA, and cell lysates. Hit identification and quantitative binding data can be generated within 90 minutes. Image Credit: Image courtesy of Adam Buckle et al., in partnership with ELRIG (UK) Ltd.
Arrayjet SMM-10: A small molecule library-on-a-chip

Figure 2. Arrayjet SMM-10 library-on-a-chip composition. Arrayjet’s 350,000 SM chemical diversity library was selected for desirable lead traits from a library of over 1 million compounds (ChemBridge). A further 100,000 SM library was selected for enhanced structural RNA binding using machine learning. A 5,000 SM sample (comprising 1,000 clusters) from both the SM diversity and RNA focussed library is miniaturized in duplicate on every Arrayjet SMM-10 screening chip. Image Credit: Image courtesy of Adam Buckle et al., in partnership with ELRIG (UK) Ltd.
RNA target binding to known small molecule hits
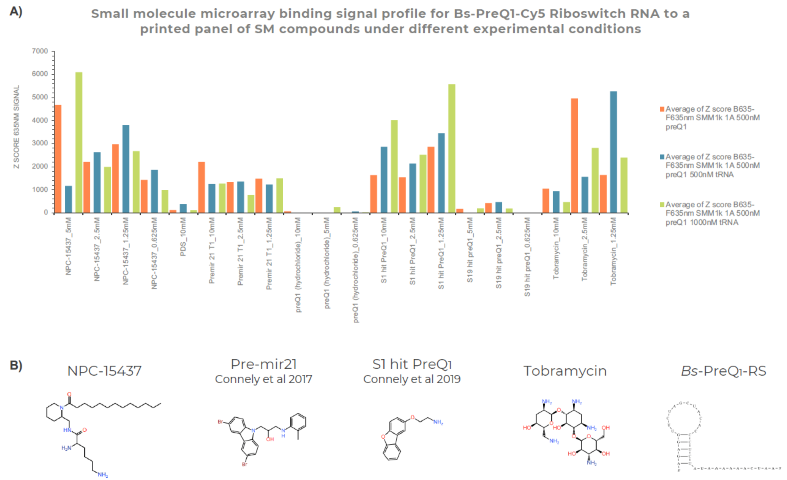
Figure 3. RNA binding signal from fluorescent SMM assay to printed and immobilized SM compounds utilizing Arrayjet’s SMM 1K format chips. A) Plot shows experimental data from three distinct RNA binding assays with 500 nM BS-PreQ1-RS, folded and screened in 40 mM Hepes, 100 mM KCL, 3 mM MgCl2, +/- additional tRNA. The X-axis shows binding for distinct printed SM compounds with known RNA binding behaviour used in Arrayjet’s SMM control panel. SM compounds include published hits from Connely et al 2017 & 2019. The y-axis is the Z-score fluorescent signal enrichment after SMM imaging with Innopsys Innoscan 710AL using F635-B635 median signal intensity to calculate the Z-score across the full 1K SMM array. B) Example SM compound structures for printed and immobilized SM compounds: NPC-15437; Pre-miR-21 SMM Target 1 (Connely et al 2017); S1 SM binding hit for BS-PreQ1-RB (Connely et al 2017); Tobramycin (a known RNA-binding aminoglycoside); and structured RNA example BS-PreQ1-RB. Image Credit: Image courtesy of Adam Buckle et al., in partnership with ELRIG (UK) Ltd.
SMM-10 screen identifies novel binders
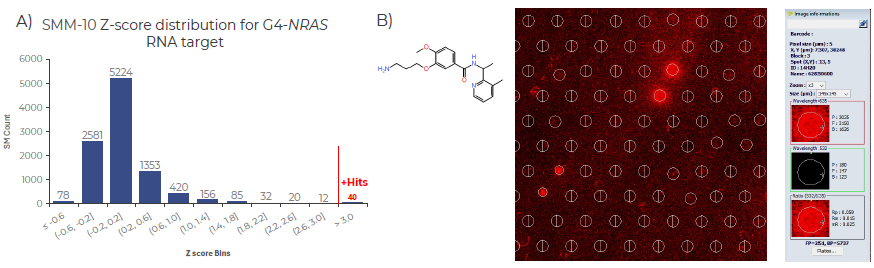
Fig 4. SMM-10 screen identifies novel SM binders for G4-NRAS RNA motif. A) Histogram illustrating the distribution of Z-score values from the SMM-10 screen using G4-NRAS-Cy5 across 10,000 printed small molecules (SMs). Z-scores are plotted along the Y-axis, with the count of SMs on the X-axis. Forty compounds with Z-scores greater than three were considered enriched binders for G4-NRAS. The RNA sequence screened corresponds to the 5′ untranslated region (UTR) of human NRAS mRNA, labelled with Cy5. B) Representative SMM hit image and chemical structure of a selected compound from the G4-NRAS screen. Image captured using Innopsys Mapix 9 software. Image Credit: Image courtesy of Adam Buckle et al., in partnership with ELRIG (UK) Ltd.
Diversity of structured RNA targets
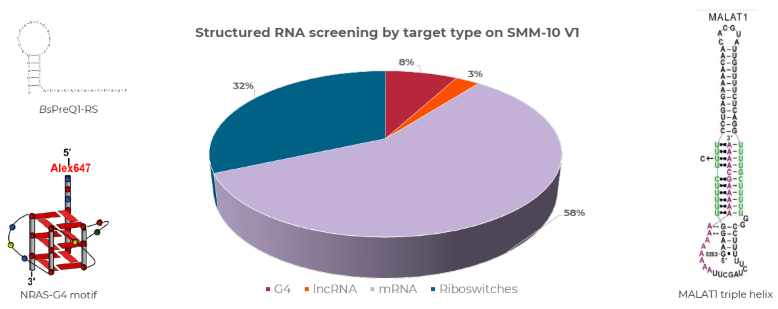
Figure 5. Percentage of SMM-10.V1 chips screened against different RNA target types by Arrayjet in 2025. RNA target types were grouped into four categories: G4 quadruplexes, long non-coding RNA (lncRNA), and mRNA riboswitches. Illustrative target structures include bacterial riboswitch Cy5-BsPreQ1-RS, Human 5’UTR NRAS-G4, and MALAT1 triple helix lncRNA fragment. Image Credit: RNA structure images from Connely et al. 2017 and Balaratnam et al. 2023. Image courtesy of Adam Buckle et al., in partnership with ELRIG (UK) Ltd.
References
- Connelly, C.M., et al. (2019). Synthetic ligands for PreQ1 riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure. Nature Communications, 10(1). DOI: 10.1038/s41467-019-09493-3. https://www.nature.com/articles/s41467-019-09493-3.
- Connelly, C.M., et al. (2017). Discovery of Inhibitors of MicroRNA-21 Processing Using Small Molecule Microarrays. ACS chemical biology, (online) 12(2), pp.435–443. DOI: 10.1021/acschembio.6b00945. https://pubs.acs.org/doi/10.1021/acschembio.6b00945.
- Bradner, J.E., et al. (2006). A method for the covalent capture and screening of diverse small molecules in a microarray format. Nature Protocols, 1(5), pp.2344–2352. DOI: 10.1038/nprot.2006.282. https://www.nature.com/articles/nprot.2006.282.
- Bradner, J.E., et al. (2006). A Robust Small-Molecule Microarray Platform for Screening Cell Lysates. Chemistry & Biology, 13(5), pp.493–504. DOI: 10.1016/j.chembiol.2006.03.004. https://linkinghub.elsevier.com/retrieve/pii/S1074552106000901
- Balaratnam, S. et al. Investigating the NRAS 5’ UTR as a target for small molecules. Cell Chem. Biol. 30, 643-657.e8 (2023).
- Abulwerdi, F.A., et al. (2019). Selective Small-Molecule Targeting of a Triple Helix Encoded by the Long Noncoding RNA, MALAT1. 14(2), pp.223–235. DOI: 10.1021/acschembio.8b00807. https://pubs.acs.org/doi/10.1021/acschembio.8b00807.
- Disney, M.D. and Seeberger, P.H. (2004). Aminoglycoside Microarrays To Explore Interactions of Antibiotics with RNAs and Proteins. Chemistry, 10(13), pp.3308–3314. DOI: 10.1002/chem.200306017. https://chemistry-europe.onlinelibrary.wiley.com/doi/10.1002/chem.200306017.
About Arrayjet Ltd
Arrayjet provides instruments and services to the pharma, diagnostic, and life science industries. Our products use inkjet technology for precision picolitre liquid handling. Arrayjet focuses on printing biological samples to create tools for genomic and proteomic screening, patient stratification, and clinical diagnosis.
The proprietary printing technology is fully automated and delivers the benefits of ease of use, precision, reproducibility, efficiency of manufacture, and total process control.
Arrayjet's patented technology simultaneously aspirates and prints multiple samples on-the-fly. This is a proven platform and its non-contact bioprinting is ideal for microarray and 96-well microplate manufacture, as well as bioprinting onto biosensors, biochips, MEMS devices, microfluidic devices, membrane sheets, or nanowell applications. Most substrates are compatible with the technology.
Arrayjet instruments offer the largest manufacturing batch size of up to 1000 slides. The instrument is modular and scalable, enabling customers to increase capacity as their requirements grow. It combines the fastest and most reliable instrumentation on the market with the versatility to print any biological sample type onto any solid substrate.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programs that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free of charge to attend!
Our values
Our values are to always ensure the highest quality of content, that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer-led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate, and collaborate on an open-access basis. We achieve this through the provision of world-class conferences, networking events, webinars, and digital content.
Sponsored Content Policy: News-Medical.Net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net, which is to educate and inform site visitors interested in medical research, science, medical devices, and treatments.
Last Updated: Dec 18, 2025