This article and associated images are based on a poster originally authored by Donato Tedesco, Tianbing Liu, Dongfang Hu, Nadya Isachenko, Debbie Deng, Paul Diehl and Alex Chenchik and presented at ELRIG Drug Discovery 2025 in affiliation with Cellecta, Inc.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Abstract
Single-cell CRISPR screening (scCRISPR) combines targeted genetic perturbations with single-cell RNA sequencing, enabling researchers to systematically link gene disruption to transcriptional outcomes at scale. This powerful approach makes it possible to interrogate hundreds to thousands of gene functions within heterogeneous cell populations from a single experiment. However, multiple scCRISPR methodologies exist, each with distinct advantages and limitations, and a clear comparison can help researchers select the optimal platform for their experimental goals.
Here, we directly compare two widely used scCRISPR partitioning technologies: droplet-based encapsulation (10x Genomics) and combinatorial split-pool barcoding (Scale Biosciences). Using four parallel screens targeting ~200 genes involved in the TNFα response, we evaluated three lentiviral vector designs (10X-CS1, CROP-seq, and a standard sgRNA vector) across both droplet-based and split-pool workflows in HEK293-Cas9 cells.
Our analysis revealed meaningful performance differences across platforms. For transcriptome coverage, the 10X-5′ and split-pool barcoding approaches captured more information per cell, with 10X-5′ recovering more mRNA molecules per cell and split-pool barcoding identifying more expressed genes. For sgRNA detection, direct capture in the 10X-5′ workflow outperformed all other methods, followed by 10X-3’, while split-pool barcoding exceeded the modified 10X-3′ protocol among indirect capture methods. Ultimately, the ability to identify drivers of the TNFα transcriptional response ranked the platforms as 10X-5′ > split-pool barcoding > modified 10X-3′ > 10X-3′.
By comparing droplet-based and split-pool scCRISPR technologies head-to-head, this study highlights the tradeoffs between transcriptome depth, sgRNA detection, and overall discovery power. These insights provide a practical guide for researchers in choosing the most appropriate methodology to match their experimental priorities, whether prioritizing high transcriptome coverage, robust perturbation detection, or flexibility in scale and cost.
Introduction
Single-cell CRISPR screens (scCRISPR)
- scCRISPR is a powerful technology for the transcriptional profiling of defined genetic perturbations. Importantly, 100s of perturbations can be assayed simultaneously in the same experiment from a single sample.
- The genetic perturbations are introduced by lentiviral transduction of sgRNA libraries in a pooled population.
- Single-cell barcoding of mRNA and sgRNA transcripts is used to uniquely label transcripts in/from each single cell.
- The identity of the genetic perturbation and the associated transcriptional response is analyzed by NGS.
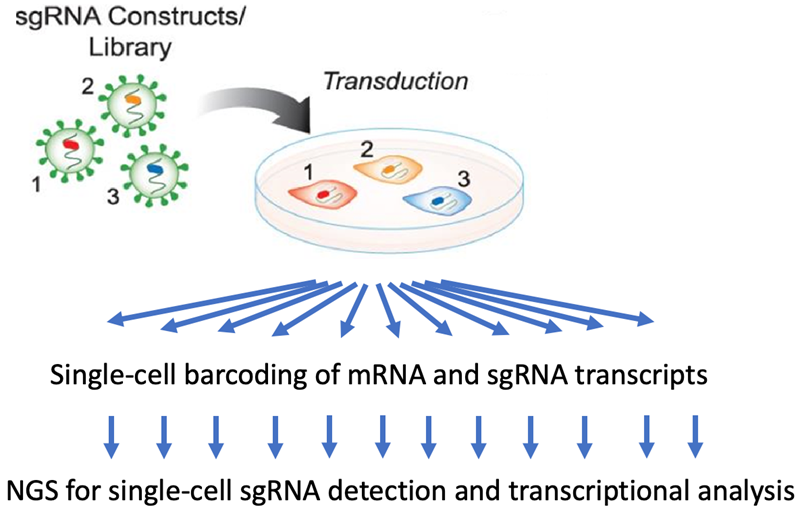
Image Credit: Image courtesy of Donato Tedesco et al., in partnership with ELRIG (UK) Ltd.
Methods
- Single-cell barcoding of transcripts can be carried out by either physical or compartment-free cell partitioning.
- Physical partitioning is carried out by mixing a cell suspension with oils to create an emulsion where micro-droplets contain single cells, which are then fused to droplets carrying beads, each carrying unique barcoded oligos for transcript barcoding.
- Compartment-free partitioning is carried out by multiple rounds of splitting-barcoding-pooling-splitting, so that each single cell receives a unique combination of multiple barcoding
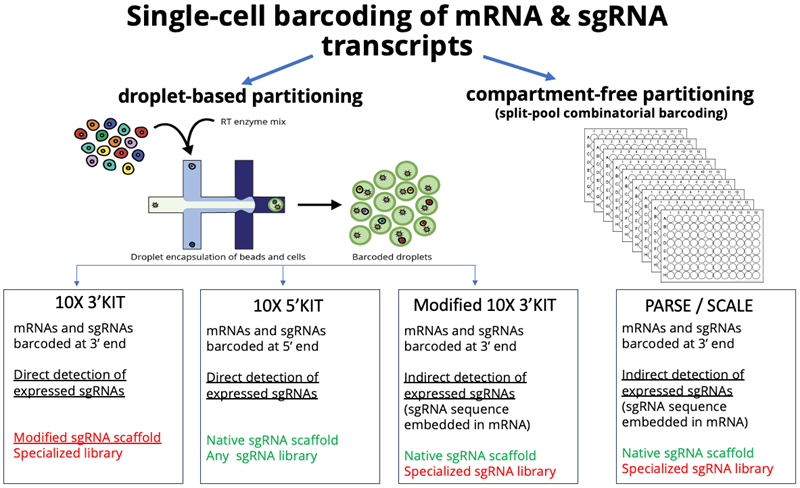
Image Credit: Image courtesy of Donato Tedesco et al., in partnership with ELRIG (UK) Ltd.
TNFa-focused sgRNA library
- 18 genes from the TNFapathway (4 sgRNAs/gene)
- 18 control genes (4 sgRNAs/gene)
- Introns + NT
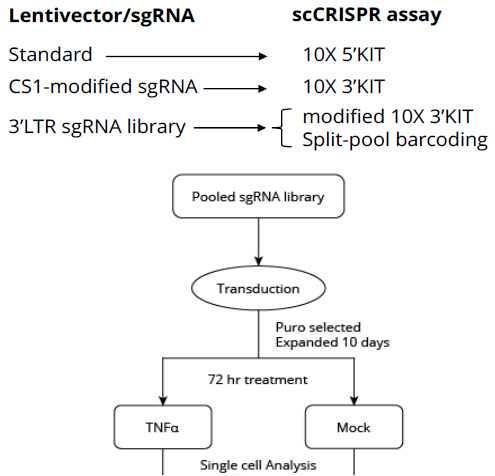
Image Credit: Image courtesy of Donato Tedesco et al., in partnership with ELRIG (UK) Ltd.
- To compare the performance of the different kits, parallel screens based on the well-characterized TNFα transcriptional response were set up.
- A small library targeting 18 genes from the TNFα pathway, 18 negative control genes + additional non-targeting controls was designed. Each gene was targeted with four different sgRNAs.
- Using Agilent oligo pools, three different libraries made from the same sgRNA set were synthesized: one for the 5’ kit, one for the 3’ kit, and one for both the 3’-modified and the combinatorial barcoding.
- The libraries were packaged, and then 293 Cas9 cells were transduced, selected, and treated as shown.
Method (cont.)
Image Credit: Image courtesy of Donato Tedesco et al., in partnership with ELRIG (UK) Ltd.
Differential expression +/- TNFα
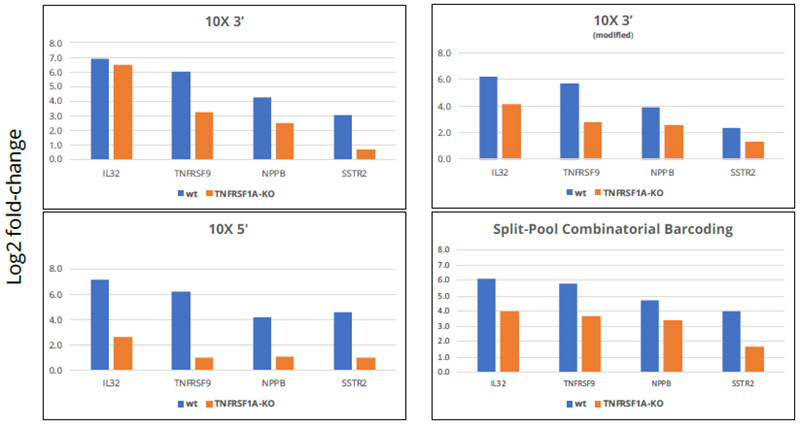
Image Credit: Image courtesy of Donato Tedesco et al., in partnership with ELRIG (UK) Ltd.
- To assess the performance of the four different methodologies, the researchers first focused on the four most up-regulated genes in response to TNFα treatment in unperturbed cells (cells expressing non-targeting sgRNAs) and examined the changes in cells that received sgRNA targeting the TNFα receptor, where the TNFα response should be disrupted.
- In all four methodologies, a disruption of the TNFα response was observed by the sgRNAs targeting the TNFα receptor. The highest level of disruption was detected with the 10X 5’ kit, where the TNFα response was abrogated entirely in three out of four cases.
Results
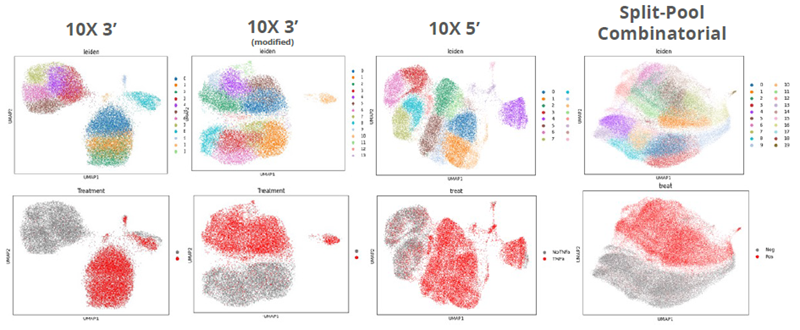
Image Credit: Image courtesy of Donato Tedesco et al., in partnership with ELRIG (UK) Ltd.
- Using Leiden clustering, the whole population of cells (treated + untreated) for each screen can be analyzed and partitioned into subgroups based on gene expression similarity. Each dot represents one cell.
- We can then figure out how TNFα-treated and untreated cells distribute across these subpopulations.
- Leiden clustering can easily distinguish between treated and untreated samples based on the transcriptional response.
Image Credit: Image courtesy of Donato Tedesco et al., in partnership with ELRIG (UK) Ltd.
- For each KO, as well as un-edited control cells, the percentage of TNFα-treated cells that clustered together with the untreated cells was calculated, indicating that the TNFα transcriptional response was inhibited.
- Most KOs and untreated cells showed very few to no treated cells with disrupted TNFα response, while this percentage of KOs showed a sizeable fraction of cells with disrupted response.
- Importantly, all seven genes happen to be critical transducers of the TNFα response.
Results (cont.)
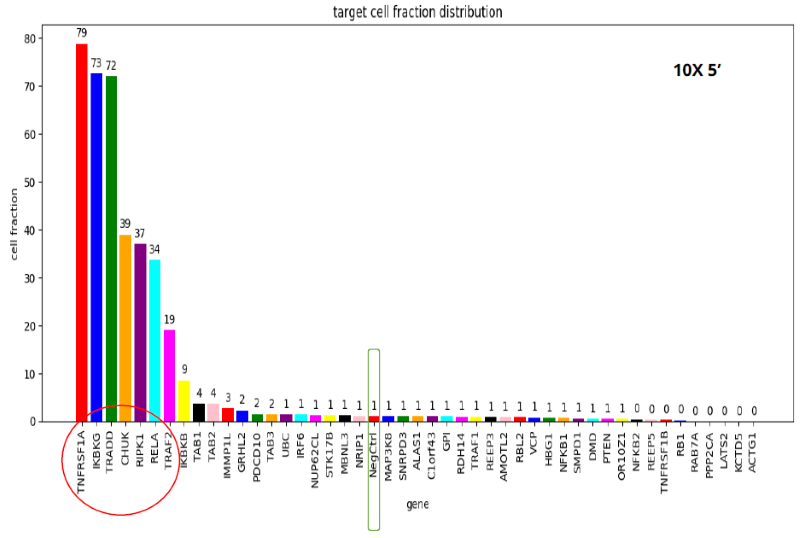
Image Credit: Image courtesy of Donato Tedesco et al., in partnership with ELRIG (UK) Ltd.
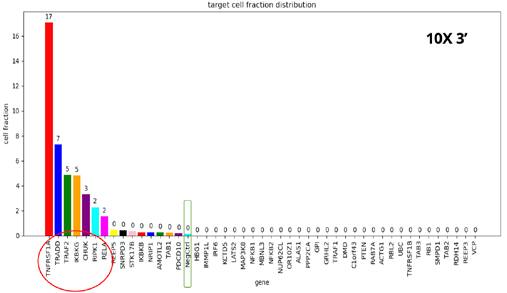
Image Credit: Image courtesy of Donato Tedesco et al., in partnership with ELRIG (UK) Ltd.
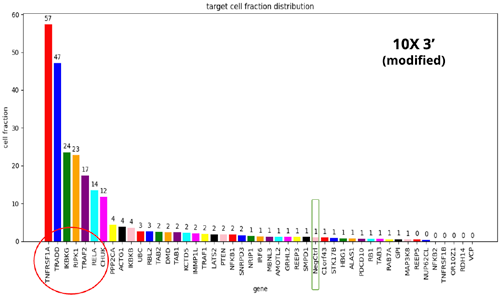
Image Credit: Image courtesy of Donato Tedesco et al., in partnership with ELRIG (UK) Ltd.
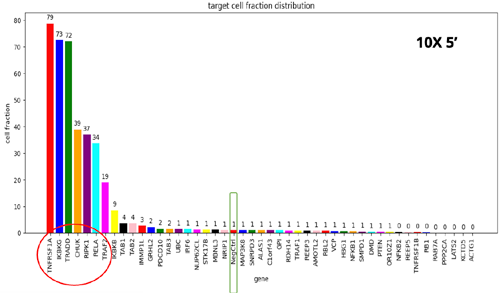
Image Credit: Image courtesy of Donato Tedesco et al., in partnership with ELRIG (UK) Ltd.
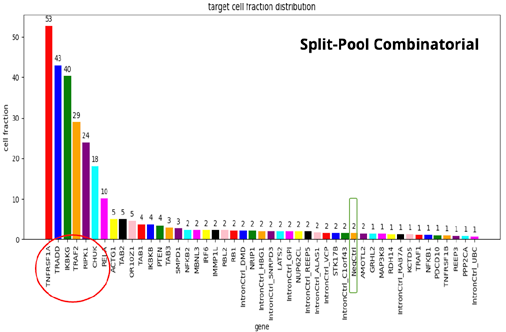
Image Credit: Image courtesy of Donato Tedesco et al., in partnership with ELRIG (UK) Ltd.
- When comparing the four different kits, we see the same set of seven gene KOs with disrupted TNFα response, but with different frequencies.
- The 10X-3’ kit is clearly the worst performer. The 3’ modified kit and the combinatorial barcoding perform equally well, but not to the level of the 10X-5’ kit.
- At this point, the researchers were curious to understand the basis of such differences and to see if there were possible ways to improve the performance of the 3’ modified and combinatorial barcoding.
- To achieve this, the researchers specifically examined the ability of each kit to detect mRNAs and sgRNA in the single-cell population.
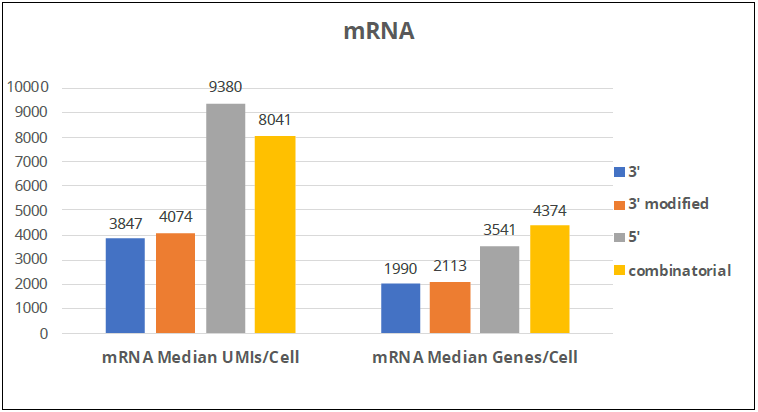
Image Credit: Image courtesy of Donato Tedesco et al., in partnership with ELRIG (UK) Ltd.
- 10X-5’ and split-pool combinatorial barcoding seem better than 10X-3’ and 10X-3’ (modified) at detecting expressed mRNAs
- Split-pool combinatorial barcoding seems to be able to detect more genes/cell
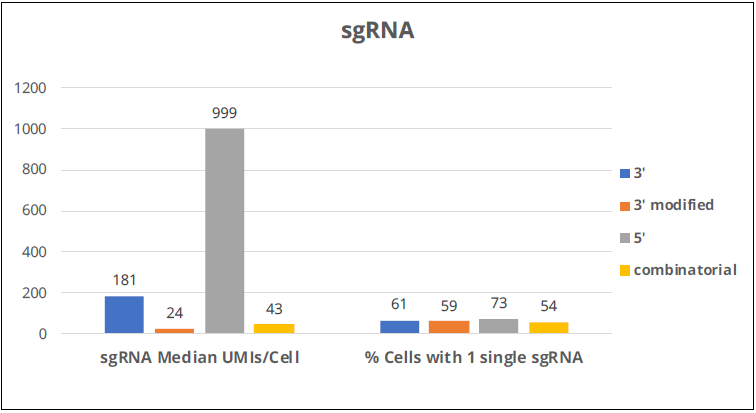
Image Credit: Image courtesy of Donato Tedesco et al., in partnership with ELRIG (UK) Ltd.
- sgRNA direct capture (10X-3’ and 10X-5’) is more efficient at detecting sgRNAs
- 10X-5’ direct capture is the most efficient, leading to the highest proportion of cells assigned to a single sgRNA
- The top histogram shows the mRNA detection in single cells, and it is clear that the 3’ kits perform worse than the 5’ kit and the combinatorial barcoding
- Interestingly, the 5’ kit seems to be the best at detecting the highest number of mRNA molecules per cell, while combinatorial barcodes seem to be able to detect the most genes per cell, which might mean that it is better than the 5’ kit at detecting low-abundance mRNAs
- Looking at sgRNA detection, it is clear that the 5’ kit is by far the best performer here, with the 3’ kit a distant second. It is essential to notice that what these two kits have in common is that they both use direct detection of sgRNAs, while modified 3’ and combinatorial barcodes use indirect detection through mRNA.
- The best performance in detecting sgRNAs translates to a higher percentage of cells with assigned single sgRNAs, so a higher percentage of cells are available for transcriptional analysis.
Discussion
- 10X 5’ appears to be the best performing protocol
- Combinatorial barcoding has some advantages over 10X 5’
- Less expensive
- Works with fixed samples
- More scalable
- Seems to detect more genes/cell
- Improved sgRNA detection by means of simple changes in the library vector structure
About Cellecta
Cellecta, Inc. was founded in April 2006 by one of the chief scientists from Clontech Laboratories. The company was borne out of the need for higher quality, more advanced shRNA and other lentiviral libraries. Our goal was to develop advanced high-throughput (HT) genetic screen technologies and their applications for the discovery and functional characterization of novel therapeutic targets and drugs.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programs that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to always ensure the highest quality of content and that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Nov 26, 2025