Introduction
Biofilms are aggregates that are formed by bacteria, which grow on surfaces. Commonly found in environments, biofilms are highly resistant to antibiotics, and as such the mechanism and regulation of biofilms are being intensively studied by scientists across the globe. This article uses a model organism, called Pseudomonas fluorescens or, to better understand the regulation of biofilm formation by this so called soil bacterium.
Genes that are involved in the formation of biofilms are often identified by transposon mutagenesis. When such a gene is injected with the transposon, the resultant mutant will exhibit reduced or increased formation of biofilms based on its function. This method was leveraged to detect a gene that is involved in the production of c-di-GMP, a bacterial second messenger.
Loss of this messenger resulted in reduced formation of biofilms in contrast to the wild type or WT strain. It was also discovered that when compared to the WT strain, the formation of biofilms was reduced significantly due to gene disruption in the de novo pathway for purine nucleotide biosynthesis. In many bacteria. It has often been reported that genes play a critical role in the pathway. As a result, these mutants were thoroughly studied to interpret the association between biofilm formation and biosynthesis pathway in P. fluorescens.
The Phenom ProX for Biofilm Research
It has been shown that mutant-induced biofilm formations were brought down to less than half of the WT strain; however, the experiments dedicated to cell counting have indicated that the total number of the mutant cells present in the biofilms were similar to that of the WT strain. To better explain this concept, it was assumed that biofilms contain mutants, whose cell size becomes smaller when compared to that of the WT strain.
To put it in simpler terms, these mutations reduce the size of the biofilm cells to a large extent. This supposed theory is not quite weird, given that the mutants are incapable of producing GMP and AMP, resulting in their slower growth. The cell sizes of the pathogens present in the biofilms can be examined through scanning electron microscopy (SEM), which is one of the best techniques available for such analysis.
As such, a desktop SEM called the Phenom ProX was used to acquire accurate data about the bacterial cell size. With the help of this instrument, a SEM image of the biofilm cells can be achieved quickly and easily following the sample setting. Thanks to this rapid process, a number of micrographs were prepared for a single strain and the same were thoroughly examined by means of the Phenom ParticleMetric software. Figure 1 depicts an example, where the Phenom ProX was used for imaging the biofilm cells of the P. fluorescens bacterium.
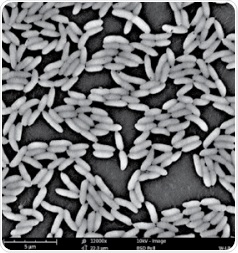
Figure 1. An example of the SEM image for the biofilm cells of P. fluorescens formed on a surface of polyvinyl chloride (magnification: ×12,000).
The Power of Phenom ParticleMetric Software
Subsequently, the SEM image of the WT strain and the SEM image of the mutants were compared which eventually showed that the mutant cells experienced the size reduction. Conversely, lack of numerical comparisons made it difficult to pinpoint the exact difference of the cell sizes between the mutants and the WT strain.
The Phenom ProX system is integrated with the ParticleMetric software, which creates a set of parameters for individual particles in SEM images. Since the P. fluorescens bacterium is rod-shaped, the limited diameter of the circle can prove to be a good estimation for the cell length. With the help of the ParticleMetric software, the confined circle diameters of over 1,500 independent cells were gathered from a single strain’s SEM images. Subsequently, histograms were produced and the medians of the limited circle diameters were measured, as shown in Figure 2.
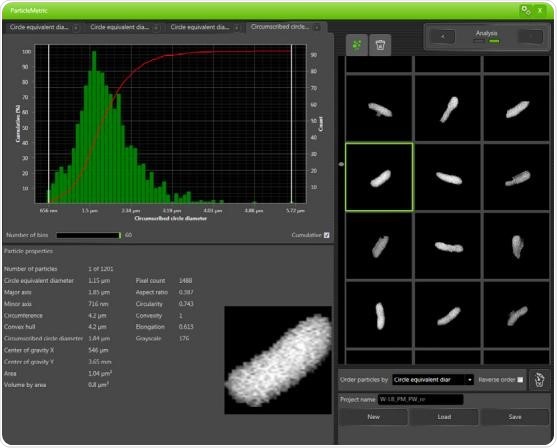
Figure 2. A histogram generated by the ParticleMetric software for the biofilm cells of P. fluorescens after collecting the circumscribed circle diameters for more than 1,500 individual cells. The median of the circumscribed circle diameter was calculated to be 1.79μm. Besides the histogram, the properties of one selected cell are being displayed.
The entire process was automatically carried out by the Phenom ParticleMetric software. However, it should be noted that the aggregated cells are sometimes treated as an individual cell when automatic particle detection is used. By using the Phenom ParticleMetric software, these aggregated cells can be intuitively eliminated to ensure reliable and consistent measurements from the data collection.
The results obtained from these measurements showed that the size and length of the mutant cells were roughly reduced by 25 to 30% when compared to the mutant cells of the WT strain. Therefore, the Phenom ParticleMetric software and the Phenom ProX desktop SEM helped in validating the theory that the size of the mutant cells present in the biofilms indeed becomes smaller when compared to that of the WT strain.
Conclusion
This article has shown how valuable data on cell sizes of bacterial biofilms can be obtained using the Phenom ProX desktop SEM in tandem with the Phenom ParticleMetric software. Given that the availability of nutrients or the presence of antibiotics govern the size and morphology of most bacteria, a better understanding of the morphology and cell size will be required for potential investigations in biofilm analyses.
Scientists studying pathogens and other microorganisms can significantly benefit from using the Phenom ProX desktop SEM and the easy-to-use ParticleMetric software. Shiro Yoshioka, Ph.D Assistant Professor Institute for Molecular Science, National Institutes of Natural Sciences Japan, said that the Phenom ProX SEM combined with the ParticleMetric software helps in measuring the cell sizes of P. fluorescens bacteria present in biofilms.
Acknowledgements
Produced from materials authored by Shiro Yoshioka from the Institute for Molecular Science, National Institutes of Natural Sciences, Japan.
About Phenom World
Phenom-World believe breakthroughs happen when complex nanotechnology is made intuitive, easier to use and brought within reach. As the leading global supplier of desktop scanning electron microscopes, their aim is to make imaging and analysis at the nanoscale available to every scientist in every lab.
That’s why Phenom-World invest their time and effort into developing high-quality electron microscope solutions that are functionally rich, yet simple to use. Because their mission is to create technology that has a real impact on how people work.
This takes innovative thinking, collaborative working and an entrepreneurial attitude. At their home in the high-tech region of Eindhoven in The Netherlands, Phenom-World have a group of passionate people who are given the freedom they need to innovate. Plus a highly skilled team of specialists in more than 40 countries who can provide support on a global scale.
Cooperation and co-creation are key to Phenom-World's success. They work closely with their partners to ensure they have the latest technologies, state-of-the-art equipment and applications for imaging and analysis. So they can get fast and accurate results and achieve more in nanotechnology.
Phenom-World is globally the yearly number 1 manufacturer of desktop scanning electron microscopes and imaging and analysis packages.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.