NanoLuc® Binary Technology, also known as NanoBiT, is an innovative binary complementation solution built on NanoLuc® Luciferase(1). As a 19 kDa enzyme, NanoLuc® Luciferase generates extremely bright, glow-type luminescence. In order to develop a complementation system, NanoLuc® Luciferase is divided into two subunits – a larger protein domain and a small peptide.
In other methods, a protein or enzyme is merely separated into two fragments. In contrast, the characteristics of each subunit are separately optimized in the NanoBiT® System. In order to achieve structural stability, the Large BiT (LgBiT; 17.6 kDa) was optimized.
The Small BiT (SmBiT; 11 amino acids) has low affinity for LgBiT and hence it was chosen for use in protein:protein interaction (PPI) assays. Both SmBiT and LgBiT are fused to the proteins of interest to detect PPIs.
Assay background is reduced by the weak affinity for self-association (KD = 190µM). When interactions occur between the two target proteins, SmBiT and LgBiT are brought close together, enabling structural complementation to produce an extremely bright, luminescent enzyme. By using the parts of the NanoBiT™ PPI Starter Systems, protein-protein interactions can be tracked in real time in living cells.
Before Protein-Protein Interactions
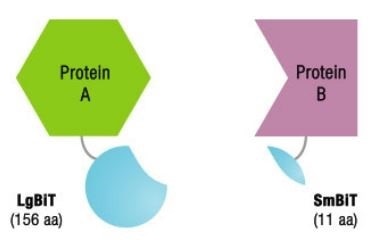
Image credit: Promega
After Protein-Protein Interactions
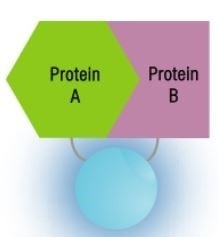
Image credit: Promega
Evaluating the NanoBiT® PPI Assay Using Two Model Systems
In this article, two PPI model systems are used to show the characteristics of the NanoBiT™ PPI Assay. The first model system is the interaction between FRB and FKBP. When rapamycin is absent, no detectable interaction occurs between FRB and FKBP. When rapamycin is present, a ternary complex is formed between rapamycin, FKBP, and FRB (2).
The second model system selected was the interaction between the subunits of protein kinase A (PKA). A PKA isoform has type 2A regulatory (PRKAR2A) and catalytic (PRKACA) subunits. These subunits interact in response to cAMP levels within the cells.
Two catalytic subunits produce an inactive complex by connecting with two regulatory subunits. When there are increasing intracellular cAMP levels, cAMP is bound to the regulatory subunit resulting in dissociation of the protein complex and discharge of catalytic subunits for downstream activity.
When there is a reduction in cAMP levels and the cAMP no longer binds to the type 2A regulatory subunit, re-association is observed with the catalytic subunit.
Comparison with A Split Firefly System
With the help of the FKBP:FRB model system, the NanoBiT® PPI Assay is compared to a split firefly luciferase system using 394–544 and 4–398 fragments (3). For each system, an optimal orientation for subunit fusion was detected, followed by using identical 10 amino acid linkers for both systems. In both methods, similar EC50 values were shown for rapamycin activation. Yet, there were key differences in the intensity of luminescence using expression under similar conditions.
NanoBiT® Luminescence at 1X Zoom
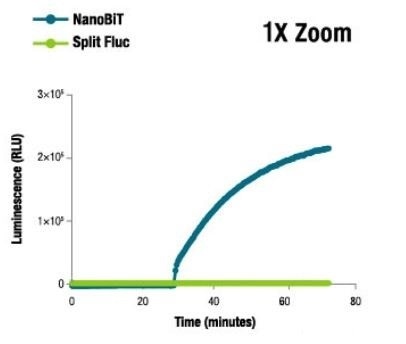
Image credit: Promega
NanoBiT® Luminescence at 1000X Zoom
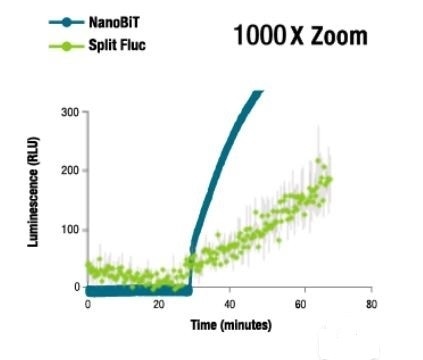
Image credit: Promega
At 37°C, NanoBiT® luminescence was found to be >1,000X brighter than that of split firefly luminescence. Due to the bright signal of the NanoBiT® PPI Assay, fusion proteins are expressed either at or near physiological levels in numerous cases that help reduce experimental artifacts.
Measuring reversible PPIs in real time in living cells
Reversibility of NanoBiT® PPI Assay in living cells was demonstrated through a PKA model system. First, expression constructs were used to transiently transfect HEK293 cells to achieve optimal orientation for fusion of SmBiT and LgBiT and the HEK293 cells were treated with a set of compounds to modulate intracellular [cAMP] levels.
As a separate measure of cAMP, a firefly luciferase-based cAMP biosensor called GloSensor™ cAMP assay was employed in replicate wells. The signal produced by the NanoBiT PPI Assay was observed to have the predicted inverse association with the cAMP GloSensor™ assay.
This demonstrates the fast signal changes in response to varying intracellular [cAMP] levels and also shows that reversible dynamics of protein interactions in living cells can be tracked using the NanoBiT™ PPI Assay.
A PKA model system
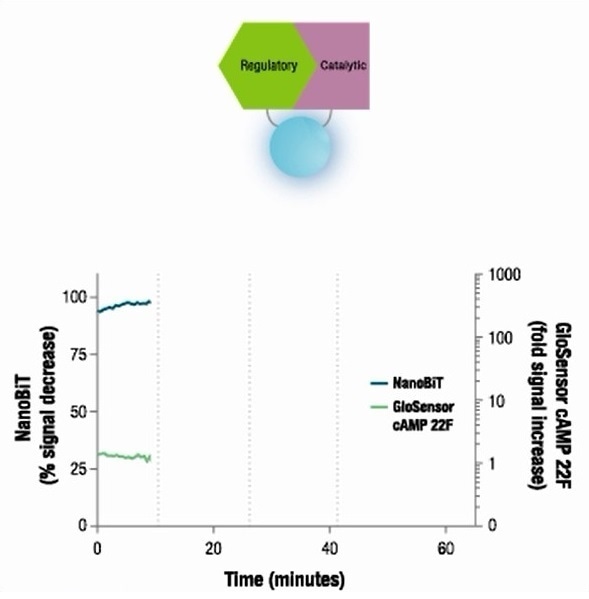
Image credit: Promega
Reversible interaction dynamics of proteins in living cells
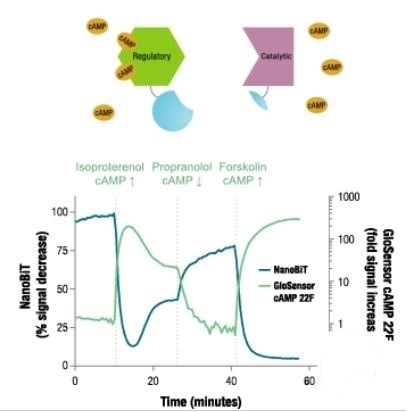
Image credit: Promega
Measuring PPIs Using Bioluminescence Imaging
With the help of the bright signal produced by the NanoBiT® PPI Assay, interactions between proteins can be observed in each cell using bioluminescence imaging. Expression constructs were used to transiently transfect HEK293 cells to achieve optimal orientation to fuse SmBiT and LgBiT to FRB and FKBP.
A saturating concentration of rapamycin was used then to treat transfected cells, and images were obtained at an interval of 30 seconds using the Olympus LV200 microscope. Individual cell response can be measured in real time, demonstrating the predicted rapid increased in signal after treatment with rapamycin.
No Signal Increase Before Rapamycin Treatment
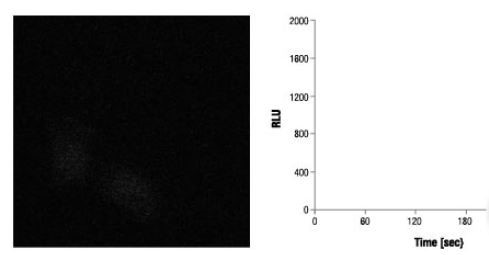
Image credit: Promega
Rapid Signal Increase After Rapamycin Treatment
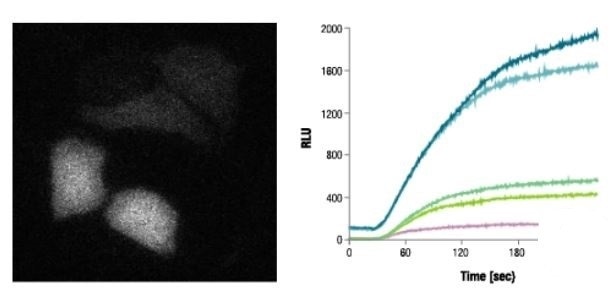
Image credit: Promega
Conclusions
As demonstrated here with two well-characterized model systems, NanoBiT® PPI technology provides a sensitive, reversible assay system that can be adapted to analysis of protein interaction dynamics throughout the cell.
References
- Dixon, A.S. et al. (2015) NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem. Biol., Articles ASAP Available from: http://pubs.acs.org/doi/abs/10.1021/acschembio.5b00753
- Banaszynski, L. A., Liu, C. W., and Wandless, T. J. (2005) Characterization of the FKBP-rapamycin-FRB ternary complex, J. Am. Chem. Soc. 127, 4715–21.
- Paulmurugan, R. and Gambhir, S.S. (2007) Combinatorial library screening for developing an improved split-firefly luciferase fragment-assisted complementation system for studying protein-protein interactions. Anal Chem. 79(6), 2346–53.
About Promega
With a portfolio of more than 3,000 products covering the fields of genomics, protein analysis and expression, cellular analysis, drug discovery and genetic identity, Promega is a global leader in providing innovative solutions and technical support to life scientists in academic, industrial and government settings.
Promega products are used by life scientists who are asking fundamental questions about biological processes as well as by scientists who are applying scientific knowledge to diagnose and treat diseases, discover new therapeutics, and use genetics and DNA testing for human identification.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and iform site visitors interested in medical research, science, medical devices and treatments.