This article and associated images are based on a poster originally authored by Marisa Amato and presented at ELRIG Drug Discovery 2025 in affiliation with Singleron Biotechnologies GmbH.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Abstract
Accurate and comprehensive RNA sequencing at the single-cell level is crucial for understanding cellular heterogeneity, gene regulation, and the complexities of disease mechanisms. Traditional short-read single-cell RNA sequencing methods often capture either the 5’ or the 3’ end of the transcripts for their quantification, which can miss critical information such as alternative splicing events, gene fusion, and allele-specific gene expression.
This limitation can obscure the true complexity of cellular transcriptomes and hinder the discovery of novel biomarkers and therapeutic targets. The research team developed a new method, MobiuSCOPE, to address these challenges by providing a full-length RNA sequencing solution that captures the entire mRNA transcript at the single-cell level.
A microwell chip, SCOPE-Chip, is used for partitioning thousands of cells into individual wells, followed by the capture and barcoding of mRNA from each single cell. An optimized reverse transcriptase (RT) formulation, which has improved enzyme processivity, is then used to generate full-length 3’ barcoded cDNA, each with a unique cell barcode and UMI next to the 3’ sequence of the mRNA.
A circularization step combined with subsequent reverse PCR is then used to bring the 5’ sequence of the same mRNA to the proximity of the cell barcode and UMI, generating a 5’ barcoded cDNA. Sequencing libraries are constructed by random fragmentation of both 3’ and 5’ cDNA pools and ligating the fragments to Illumina sequencing adaptors.
All reads coming from the same transcript have the same cell barcode and UMI and can be assembled by a proprietary bioinformatics pipeline to yield full-length transcript information. The MobiuSCOPE technology combines advanced molecular barcoding technology with a highly sensitive workflow that minimizes technical biases, ensuring precise and reproducible results.
It is scalable, supporting the analysis of hundreds to thousands of single cells, making it suitable for a wide range of applications, including the detection and characterization of splicing isoforms, fusion genes, expressed mutations, or SNPs under diverse physiological conditions at the single-cell level.
Technical principle
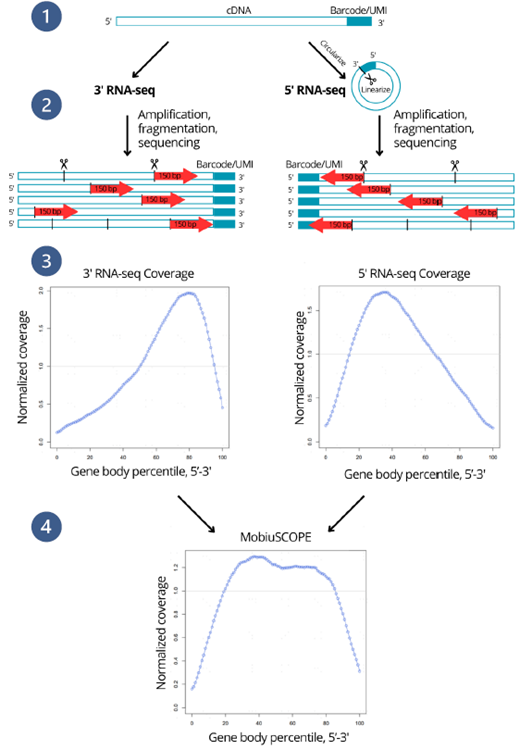
Figure 1. Technical principles of MobiuSCOPE.
1. cDNA generation. Left: 3’ barcoded cDNA is generated by adding a unique cell barcode and UMI to the 3’ end; Right: 5’ barcoded cDNA is generated by circularizing the 3’ barcoded cDNA, bringing the 5’ end close to the cell barcode and UMI. The cDNA is then linearized through PCR, generating a 5’ barcoded cDNA
2. Library construction. Sequencing libraries are constructed by random fragmentation of both 3’ and 5’ cDNA pools. Red arrows indicate the coverage of PE150 sequencing reads of individual fragments.
3. Sequencing. The 3’ and 5’ libraries are sequenced. Left: the 3’ library is biased towards the 3’ end. Right: the 5’ library is biased towards the 5’ end.
4. 3’ and 5’ library assembly. MobiuSCOPE combines the 3’ and 5’ libraries to achieve full-length coverage. All reads coming from the same transcript have the same cell barcode and UMI and can be assembled by a proprietary bioinformatics pipeline to yield full-length transcript information. Image Credit: Image courtesy of Marisa Amato, in partnership with ELRIG (UK) Ltd.
MobiuSCOPE provides full gene body coverage
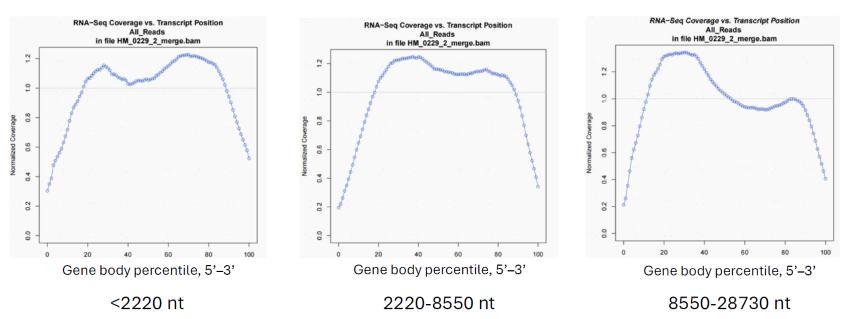
Figure 2. MobiuSCOPE provides full gene body coverage of up to 28730 nucleotides. Plots of MobiuSCOPE RNA-sequencing coverage versus transcript position of transcripts of different lengths. Y-axis: normalized coverage. X-axis: transcript position in terms of gene body percentile, where 0 % indicates the 5’ end, and 100 % indicates the 3’ end.
Left: For transcripts of up to 2220 nucleotides (nt), MobiuSCOPE showed even gene body coverage from 5’ to 3’ end. Middle: Similarly, for transcripts ranging from 2220 to 8550 nt, MobiuSCOPE is able to cover the entire transcript. Right: For large transcripts with more than 8550 nt, MobiuSCOPE still shows full gene body coverage, with a dip in coverage in the middle of the transcript. Image Credit: Image courtesy of Marisa Amato, in partnership with ELRIG (UK) Ltd.
MobiuSCOPE detects more splice junctions compared to 3‘ or 5‘ RNA sequencing
Table 1. Sequencing metrics comparison between 3’ single cell RNA sequencing (scRNA-seq), 5’ scRNA-seq, and MobiuSCOPE. RNA splicing is an important step in the eukaryotic gene expression process that contributes to the functional diversity of the transcriptome. Aberrant regulation of the splicing events are often associated with diseases. Source: ELRIG (UK) Ltd.
|
3’ RNA sequencing |
5’ RNA sequencing |
MobiuSCOPE |
| Cell number |
5737 |
6027 |
6384 |
| Median UMIs per cell |
4046 |
5062 |
7644 |
| Number of slice junctions (SJ) |
318,607 |
316,785 |
382,485 |
| Median SJs per cell |
1984 |
2223 |
3349 |
| Median reads per SJ |
30 |
33 |
61 |
Mixtures of human PBMCs and mouse spleen cells were analyzed using either 3‘scRNA-seq, 5‘scRNA-seq, or MobiuSCOPE. All methods identified similar cell numbers, with MobiuSCOPE detecting 11 % and 3 % higher numbers of cells compared to 3’ and 5’ scRNA-seq, respectively.
MobiuSCOPE identified a significantly higher number of splice junctions (20 % and 21 % higher than 3’ and 5’ scRNA-seq, respectively). The median splice junctions per cell were 69 % and 51 % higher than 3’ and 5’ scRNA-seq, respectively. The median reads per splice junction by MobiuSCOPE were almost double that of 3’ and 5’ scRNA-seq.
MobiuSCOPE detects fusion genes with high sensitivity
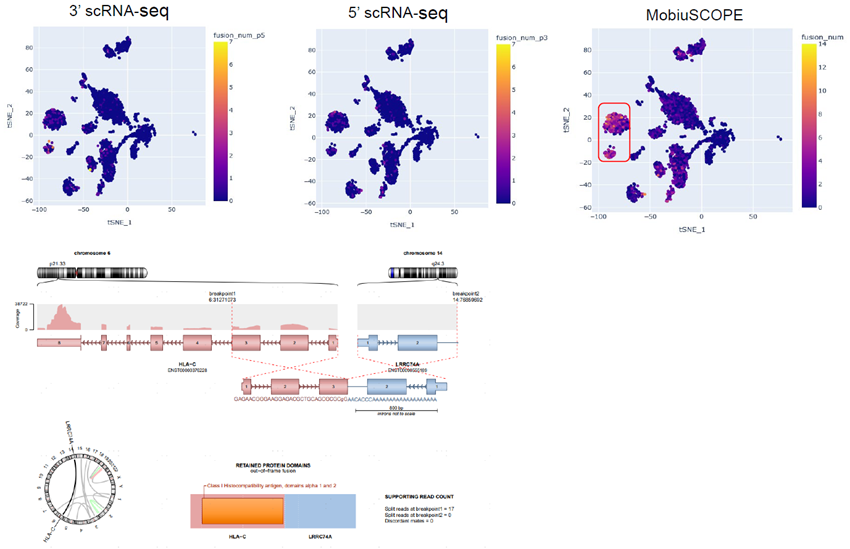
Figure 3. MobiuSCOPE detected HLA-C_LRRC74A gene fusion with high sensitivity. A PBMC sample from a patient was processed with the MobiuSCOPE method to generate full-length single-cell RNA sequencing data.
Top: A gene fusion of HLA-C and LRRC74A was identified and shown to be expressed at different levels in different cell types. MobiuSCOPE detected a significantly higher level of the fusion gene compared to 3’ or 5’ scRNA-seq alone. Bottom: Visualization of HCA-C_LRRC74A gene fusion using Arriba. Image Credit: Image courtesy of Marisa Amato, in partnership with ELRIG (UK) Ltd.
Conclusion
The MobiuSCOPE single-cell full-length RNA library construction method offers a straightforward approach for constructing single-cell libraries, utilizing short-read sequencing to generate full-length RNA information at single-cell resolution.
The accuracy and throughput of short-read sequencing enable the detection of splice junctions, fusion genes, mutations, and allele-specific expression from thousands of genes in each cell, as well as from tens of thousands of cells in parallel.
About Singleron
Singleron delivers single cell sequencing solutions to researchers and clinicians worldwide. With offices and laboratories in Germany, Singapore, North America, and China, the company provides advanced kits and sequencing services designed to unlock cellular insights. Singleron’s commitment to innovation and precision drives the development of cutting-edge technologies that empower scientific discovery.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programmes that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to always ensure the highest quality of content and that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Nov 13, 2025